Chapter 4 Pandas
4.1 Introduction
pandas is the Python Data Analysis package. It allows for data ingestion, transformation and cleaning, and creates objects that can then be passed on to analytic packages like statsmodels and scikit-learn for modeling and packages like matplotlib, seaborn, and plotly for visualization.
pandas is built on top of numpy, so many numpy functions are commonly used in manipulating pandas objects.
pandasis a pretty extensive package, and we’ll only be able to cover some of its features. For more details, there is free online documentation at pandas.pydata.org. You can also look at the book “Python for Data Analysis (2nd edition)” by Wes McKinney, the original developer of the pandas package, for more details.
4.2 Starting pandas
As with any Python module, you have to “activate” pandas by using import. The “standard” alias for pandas is pd. We will also import numpy, since pandas uses some numpy functions in the workflows.
4.3 Data import and export
Most data sets you will work with are set up in tables, so are rectangular in shape. Think Excel spreadsheets. In pandas the structure that will hold this kind of data is a DataFrame. We can read external data into a DataFrame using one of many read_* functions. We can also write from a DataFrame to a variety of formats using to_* functions. The most common of these are listed below:
| Format type | Description | reader | writer |
|---|---|---|---|
| text | CSV | read_csv | to_csv |
| Excel | read_excel | to_excel | |
| text | JSON | read_json | to_json |
| binary | Feather | read_feather | to_feather |
| binary | SAS | read_sas | |
| SQL | SQL | read_sql | to_sql |
We’ll start by reading in the mtcars dataset stored as a CSV file
make mpg cyl disp hp ... qsec vs am gear carb
0 Mazda RX4 21.0 6 160.0 110 ... 16.46 0 1 4 4
1 Mazda RX4 Wag 21.0 6 160.0 110 ... 17.02 0 1 4 4
2 Datsun 710 22.8 4 108.0 93 ... 18.61 1 1 4 1
3 Hornet 4 Drive 21.4 6 258.0 110 ... 19.44 1 0 3 1
4 Hornet Sportabout 18.7 8 360.0 175 ... 17.02 0 0 3 2
5 Valiant 18.1 6 225.0 105 ... 20.22 1 0 3 1
6 Duster 360 14.3 8 360.0 245 ... 15.84 0 0 3 4
7 Merc 240D 24.4 4 146.7 62 ... 20.00 1 0 4 2
8 Merc 230 22.8 4 140.8 95 ... 22.90 1 0 4 2
9 Merc 280 19.2 6 167.6 123 ... 18.30 1 0 4 4
10 Merc 280C 17.8 6 167.6 123 ... 18.90 1 0 4 4
11 Merc 450SE 16.4 8 275.8 180 ... 17.40 0 0 3 3
12 Merc 450SL 17.3 8 275.8 180 ... 17.60 0 0 3 3
13 Merc 450SLC 15.2 8 275.8 180 ... 18.00 0 0 3 3
14 Cadillac Fleetwood 10.4 8 472.0 205 ... 17.98 0 0 3 4
15 Lincoln Continental 10.4 8 460.0 215 ... 17.82 0 0 3 4
16 Chrysler Imperial 14.7 8 440.0 230 ... 17.42 0 0 3 4
17 Fiat 128 32.4 4 78.7 66 ... 19.47 1 1 4 1
18 Honda Civic 30.4 4 75.7 52 ... 18.52 1 1 4 2
19 Toyota Corolla 33.9 4 71.1 65 ... 19.90 1 1 4 1
20 Toyota Corona 21.5 4 120.1 97 ... 20.01 1 0 3 1
21 Dodge Challenger 15.5 8 318.0 150 ... 16.87 0 0 3 2
22 AMC Javelin 15.2 8 304.0 150 ... 17.30 0 0 3 2
23 Camaro Z28 13.3 8 350.0 245 ... 15.41 0 0 3 4
24 Pontiac Firebird 19.2 8 400.0 175 ... 17.05 0 0 3 2
25 Fiat X1-9 27.3 4 79.0 66 ... 18.90 1 1 4 1
26 Porsche 914-2 26.0 4 120.3 91 ... 16.70 0 1 5 2
27 Lotus Europa 30.4 4 95.1 113 ... 16.90 1 1 5 2
28 Ford Pantera L 15.8 8 351.0 264 ... 14.50 0 1 5 4
29 Ferrari Dino 19.7 6 145.0 175 ... 15.50 0 1 5 6
30 Maserati Bora 15.0 8 301.0 335 ... 14.60 0 1 5 8
31 Volvo 142E 21.4 4 121.0 109 ... 18.60 1 1 4 2
[32 rows x 12 columns]This just prints out the data, but then it’s lost. To use this data, we have to give it a name, so it’s stored in Python’s memory
One of the big differences between a spreadsheet program and a programming language from the data science perspective is that you have to load data into the programming language. It’s not “just there” like Excel. This is a good thing, since it allows the common functionality of the programming language to work across multiple data sets, and also keeps the original data set pristine. Excel users can run into problems and corrupt their data if they are not careful.
If we wanted to write this data set back out into an Excel file, say, we could do
You may get an error if you don’t have the
openpyxlpackage installed. You can easily install it from the Anaconda prompt usingconda install openpyxland following the prompts.
4.4 Exploring a data set
We would like to get some idea about this data set. There are a bunch of functions linked to the DataFrame object that help us in this. First we will use head to see the first 8 rows of this data set
make mpg cyl disp hp ... qsec vs am gear carb
0 Mazda RX4 21.0 6 160.0 110 ... 16.46 0 1 4 4
1 Mazda RX4 Wag 21.0 6 160.0 110 ... 17.02 0 1 4 4
2 Datsun 710 22.8 4 108.0 93 ... 18.61 1 1 4 1
3 Hornet 4 Drive 21.4 6 258.0 110 ... 19.44 1 0 3 1
4 Hornet Sportabout 18.7 8 360.0 175 ... 17.02 0 0 3 2
5 Valiant 18.1 6 225.0 105 ... 20.22 1 0 3 1
6 Duster 360 14.3 8 360.0 245 ... 15.84 0 0 3 4
7 Merc 240D 24.4 4 146.7 62 ... 20.00 1 0 4 2
[8 rows x 12 columns]This is our first look into this data. We notice a few things. Each column has a name, and each row has an index, starting at 0.
If you’re interested in the last N rows, there is a corresponding
tailfunction
Let’s look at the data types of each of the columns
make object
mpg float64
cyl int64
disp float64
hp int64
drat float64
wt float64
qsec float64
vs int64
am int64
gear int64
carb int64
dtype: objectThis tells us that some of the variables, like mpg and disp, are floating point (decimal) numbers, several are integers, and make is an “object”. The dtypes function borrows from numpy, where there isn’t really a type for character or categorical variables. So most often, when you see “object” in the output of dtypes, you think it’s a character or categorical variable.
We can also look at the data structure in a bit more detail.
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 32 entries, 0 to 31
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 make 32 non-null object
1 mpg 32 non-null float64
2 cyl 32 non-null int64
3 disp 32 non-null float64
4 hp 32 non-null int64
5 drat 32 non-null float64
6 wt 32 non-null float64
7 qsec 32 non-null float64
8 vs 32 non-null int64
9 am 32 non-null int64
10 gear 32 non-null int64
11 carb 32 non-null int64
dtypes: float64(5), int64(6), object(1)
memory usage: 3.1+ KBThis tells us that this is indeed a DataFrame, wth 12 columns, each with 32 valid observations. Each row has an index value ranging from 0 to 11. We also get the approximate size of this object in memory.
You can also quickly find the number of rows and columns of a data set by using shape, which is borrowed from numpy.
(32, 12)More generally, we can get a summary of each variable using the describe function
mpg cyl disp ... am gear carb
count 32.000000 32.000000 32.000000 ... 32.000000 32.000000 32.0000
mean 20.090625 6.187500 230.721875 ... 0.406250 3.687500 2.8125
std 6.026948 1.785922 123.938694 ... 0.498991 0.737804 1.6152
min 10.400000 4.000000 71.100000 ... 0.000000 3.000000 1.0000
25% 15.425000 4.000000 120.825000 ... 0.000000 3.000000 2.0000
50% 19.200000 6.000000 196.300000 ... 0.000000 4.000000 2.0000
75% 22.800000 8.000000 326.000000 ... 1.000000 4.000000 4.0000
max 33.900000 8.000000 472.000000 ... 1.000000 5.000000 8.0000
[8 rows x 11 columns]These are usually the first steps in exploring the data.
4.5 Data structures and types
pandas has two main data types: Series and DataFrame. These are analogous to vectors and matrices, in that a Series is 1-dimensional while a DataFrame is 2-dimensional.
4.5.1 pandas.Series
The Series object holds data from a single input variable, and is required, much like numpy arrays, to be homogeneous in type. You can create Series objects from lists or numpy arrays quite easily
0 1.0
1 3.0
2 5.0
3 NaN
4 9.0
5 13.0
dtype: float640 1
1 2
2 3
3 4
4 5
5 6
6 7
7 8
8 9
9 10
10 11
11 12
12 13
13 14
14 15
15 16
16 17
17 18
18 19
dtype: int64You can access elements of a Series much like a dict
5There is no requirement that the index of a Series has to be numeric. It can be any kind of scalar object
a -0.283473
b 0.157530
c 1.051739
d 0.859905
e 1.178951
dtype: float640.859904696094078a -0.283473
b 0.157530
c 1.051739
d 0.859905
dtype: float64Well, slicing worked, but it gave us something different than expected. It gave us both the start and end of the slice, which is unlike what we’ve encountered so far!!
It turns out that in pandas, slicing by index actually does this. It is a discrepancy from numpy and Python in general that we have to be careful about.
You can extract the actual values into a numpy array
array([-0.28347282, 0.1575304 , 1.05173885, 0.8599047 , 1.17895111])In fact, you’ll see that much of pandas’ structures are build on top of numpy arrays. This is a good thing, since you can take advantage of the powerful numpy functions that are built for fast, efficient scientific computing.
Making the point about slicing again,
array([-0.28347282, 0.1575304 , 1.05173885])This is different from index-based slicing done earlier.
4.5.2 pandas.DataFrame
The DataFrame object holds a rectangular data set. Each column of a DataFrame is a Series object. This means that each column of a DataFrame must be comprised of data of the same type, but different columns can hold data of different types. This structure is extremely useful in practical data science. The invention of this structure was, in my opinion, transformative in making Python an effective data science tool.
4.5.2.1 Creating a DataFrame
The DataFrame can be created by importing data, as we saw in the previous section. It can also be created by a few methods within Python.
First, it can be created from a 2-dimensional numpy array.
0 1 2 3 4
0 0.228273 1.026890 -0.839585 -0.591182 -0.956888
1 -0.222326 -0.619915 1.837905 -2.053231 0.868583
2 -0.920734 -0.232312 2.152957 -1.334661 0.076380
3 -1.246089 1.202272 -1.049942 1.056610 -0.419678You will notice that it creates default column names, that are merely the column number, starting from 0. We can also create the column names and row index (similar to the Series index we saw earlier) directly during creation.
d2 = pd.DataFrame(rng.normal(0,1, (4, 5)),
columns = ['A','B','C','D','E'],
index = ['a','b','c','d'])
d2 A B C D E
a 2.294842 -2.594487 2.822756 0.680889 -1.577693
b -1.976254 0.533340 -0.290870 -0.513520 1.982626
c 0.226001 -1.839905 1.607671 0.388292 0.399732
d 0.405477 0.217002 -0.633439 0.246622 -1.939546We could also create a
DataFramefrom a list of lists, as long as things line up, just as we showed fornumpyarrays. However, to me, other ways, including thedictmethod below, make more sense.
We can change the column names (which can be extracted and replaced with the columns attribute) and the index values (using the index attribute).
Index(['A', 'B', 'C', 'D', 'E'], dtype='object')d2.columns = pd.Index(['V'+str(i) for i in range(1,6)]) # Index creates the right objects for both column names and row names, which can be extracted and changed with the `index` attribute
d2 V1 V2 V3 V4 V5
a 2.294842 -2.594487 2.822756 0.680889 -1.577693
b -1.976254 0.533340 -0.290870 -0.513520 1.982626
c 0.226001 -1.839905 1.607671 0.388292 0.399732
d 0.405477 0.217002 -0.633439 0.246622 -1.939546Exercise: Can you explain what I did in the list comprehension above? The key points are understanding str and how I constructed the range.
V1 V2 V3 V4 V5
o1 2.294842 -2.594487 2.822756 0.680889 -1.577693
o2 -1.976254 0.533340 -0.290870 -0.513520 1.982626
o3 0.226001 -1.839905 1.607671 0.388292 0.399732
o4 0.405477 0.217002 -0.633439 0.246622 -1.939546You can also extract data from a homogeneous DataFrame to a numpy array
array([[ 0.22827309, 1.0268903 , -0.83958485, -0.59118152, -0.9568883 ],
[-0.22232569, -0.61991511, 1.83790458, -2.05323076, 0.86858305],
[-0.92073444, -0.23231186, 2.1529569 , -1.33466147, 0.07637965],
[-1.24608928, 1.20227231, -1.04994158, 1.05661011, -0.41967767]])It turns out that you can use
to_numpyfor a non-homogeneousDataFrameas well.numpyjust makes it homogeneous by assigning each column the data typeobject. This also limits what you can do innumpywith the array and may require changing data types using theastypefunction. There is some more detail about theobjectdata type in the Python Tools for Data Science (notebook, PDF) document.
The other easy way to create a DataFrame is from a dict object, where each component object is either a list or a numpy array, and is homogeneous in type. One exception is if a component is of size 1; then it is repeated to meet the needs of the DataFrame’s dimensions
df = pd.DataFrame({
'A':3.,
'B':rng.random_sample(5),
'C': pd.Timestamp('20200512'),
'D': np.array([6] * 5),
'E': pd.Categorical(['yes','no','no','yes','no']),
'F': 'NIH'})
df A B C D E F
0 3.0 0.958092 2020-05-12 6 yes NIH
1 3.0 0.883201 2020-05-12 6 no NIH
2 3.0 0.295432 2020-05-12 6 no NIH
3 3.0 0.512376 2020-05-12 6 yes NIH
4 3.0 0.088702 2020-05-12 6 no NIH<class 'pandas.core.frame.DataFrame'>
RangeIndex: 5 entries, 0 to 4
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 A 5 non-null float64
1 B 5 non-null float64
2 C 5 non-null datetime64[ns]
3 D 5 non-null int64
4 E 5 non-null category
5 F 5 non-null object
dtypes: category(1), datetime64[ns](1), float64(2), int64(1), object(1)
memory usage: 429.0+ bytesWe note that C is a date object, E is a category object, and F is a text/string object. pandas has excellent time series capabilities (having origins in FinTech), and the TimeStamp function creates datetime objects which can be queried and manipulated in Python. We’ll describe category data in the next section.
You can also create a DataFrame where each column is composed of composite objects, like lists and dicts, as well. This might have limited value in some settings, but may be useful in others. In particular, this allows capabilities like the list-column construct in R tibbles. For example,
pd.DataFrame({'list' :[[1,2],[3,4],[5,6]],
'tuple' : [('a','b'), ('c','d'), ('e','f')],
'set' : [{'A','B','C'}, {'D','E'}, {'F'}],
'dicts' : [{'A': [1,2,3]}, {'B':[5,6,8]}, {'C': [3,9]}]}) list tuple set dicts
0 [1, 2] (a, b) {B, A, C} {'A': [1, 2, 3]}
1 [3, 4] (c, d) {E, D} {'B': [5, 6, 8]}
2 [5, 6] (e, f) {F} {'C': [3, 9]}4.5.2.2 Working with a DataFrame
You can extract particular columns of a DataFrame by name
0 yes
1 no
2 no
3 yes
4 no
Name: E, dtype: category
Categories (2, object): [no, yes]0 0.958092
1 0.883201
2 0.295432
3 0.512376
4 0.088702
Name: B, dtype: float64There is also a shortcut for accessing single columns, using Python’s dot (
.) notation.
0 0.958092
1 0.883201
2 0.295432
3 0.512376
4 0.088702
Name: B, dtype: float64This notation can be more convenient if we need to perform operations on a single column. If we want to extract multiple columns, this notation will not work. Also, if we want to create new columns or replace existing columns, we need to use the array notation with the column name in quotes.
Let’s look at slicing a DataFrame
4.5.2.3 Extracting rows and columns
There are two extractor functions in pandas:
locextracts by label (index label, column label, slice of labels, etc.ilocextracts by index (integers, slice objects, etc.
df2 = pd.DataFrame(rng.randint(0,10, (5,4)),
index = ['a','b','c','d','e'],
columns = ['one','two','three','four'])
df2 one two three four
a 5 3 2 8
b 9 3 0 5
c 8 4 3 3
d 5 2 7 1
e 6 7 8 7First, let’s see what naively slicing this DataFrame does.
a 5
b 9
c 8
d 5
e 6
Name: one, dtype: int64Ok, that works. It grabs one column from the dataset. How about the dot notation?
a 5
b 9
c 8
d 5
e 6
Name: one, dtype: int64Let’s see what this produces.
<class 'pandas.core.series.Series'>So this is a series, so we can potentially do slicing of this series.
9b 9
c 8
d 5
Name: one, dtype: int64a 5
b 9
c 8
Name: one, dtype: int64Ok, so we have all the Series slicing available. The problem here is in semantics, in that we are grabbing one column and then slicing the rows. That doesn’t quite work with our sense that a DataFrame is a rectangle with rows and columns, and we tend to think of rows, then columns.
Let’s see if we can do column slicing with this.
one two three four
a 5 3 2 8
b 9 3 0 5
c 8 4 3 3
d 5 2 7 1
e 6 7 8 7That’s not what we want, of course. It’s giving back the entire data frame. We’ll come back to this.
one three
a 5 2
b 9 0
c 8 3
d 5 7
e 6 8That works correctly though. We can give a list of column names. Ok.
How about row slices?
one two three four
a 5 3 2 8
b 9 3 0 5
c 8 4 3 3Ok, that works. It slices rows, but includes the largest index, like a Series but unlike numpy arrays.
one two three four
a 5 3 2 8
b 9 3 0 5Slices by location work too, but use the numpy slicing rules.
This entire extraction method becomes confusing. Let’s simplify things for this, and then move on to more consistent ways to extract elements of a DataFrame. Let’s agree on two things. If we’re going the direct extraction route,
- We will extract single columns of a
DataFramewith[]or., i.e.,df2['one']ordf2.one - We will extract slices of rows of a
DataFrameusing location only, i.e.,df2[:3].
For everything else, we’ll use two functions, loc and iloc.
locextracts elements like a matrix, using index and columnsilocextracts elements like a matrix, using location
one two three
a 5 3 2
b 9 3 0
c 8 4 3
d 5 2 7
e 6 7 8 one two three four
a 5 3 2 8
b 9 3 0 5
c 8 4 3 3
d 5 2 7 10So loc works just like a matrix, but with pandas slicing rules (include largest index)
two three four
a 3 2 8
b 3 0 5
c 4 3 3
d 2 7 1
e 7 8 7 one two three four
b 9 3 0 5
c 8 4 3 3 two three four
b 3 0 5
c 4 3 3iloc slices like a matrix, but uses numpy slicing conventions (does not include highest index)
If we want to extract a single element from a dataset, there are two functions available, iat and at, with behavior corresponding to iloc and loc, respectively.
304.5.2.4 Boolean selection
We can also use tests to extract data from a DataFrame. For example, we can extract only rows where column labeled one is greater than 3.
one two three four
a 5 3 2 8
b 9 3 0 5
c 8 4 3 3
d 5 2 7 1
e 6 7 8 7We can also do composite tests. Here we ask for rows where one is greater than 3 and three is less than 9
one two three four
a 5 3 2 8
b 9 3 0 5
c 8 4 3 3
d 5 2 7 1
e 6 7 8 74.5.2.5 query
DataFrame’s have a query method allowing selection using a Python expression
a b c
0 0.824319 0.095623 0.459736
1 0.200426 0.302179 0.534959
2 0.381447 0.775250 0.654027
3 0.482755 0.512036 0.763493
4 0.679697 0.972007 0.946838
5 0.111028 0.909652 0.136886
6 0.817464 0.499767 0.769067
7 0.921952 0.481407 0.479161
8 0.626052 0.484680 0.518859
9 0.497046 0.045412 0.324195 a b c
1 0.200426 0.302179 0.534959
3 0.482755 0.512036 0.763493We can equivalently write this query as
a b c
1 0.200426 0.302179 0.534959
3 0.482755 0.512036 0.7634934.5.2.6 Replacing values in a DataFrame
We can replace values within a DataFrame either by position or using a query.
one two three four
a 5 3 2 8
b 9 3 0 5
c 8 4 3 3
d 5 2 7 1
e 6 7 8 7 one two three four
a 2 3 2 8
b 5 3 0 5
c 2 4 3 3
d 5 2 7 1
e 2 7 8 7 one two three four
a 2 3 2 8
b 5 3 0 5
c 2 4 3 -9
d 5 2 7 1
e 2 7 8 7Let’s now replace values using replace which is more flexible.
one two three four
a 2 3 2 8
b 5 3 -9 5
c 2 4 3 -9
d 5 2 7 1
e 2 7 8 7 one two three four
a 2.5 3.0 2.5 6.5
b 5.0 3.0 0.0 5.0
c 2.5 4.0 3.0 -9.0
d 5.0 2.5 7.0 1.0
e 2.5 7.0 6.5 7.0df2.replace({'one': {5: 500}, 'three': {0: -9, 8: 800}})
# different replacements in different columns one two three four
a 2 3 2 8
b 500 3 -9 5
c 2 4 3 -9
d 500 2 7 1
e 2 7 800 7See more examples in the documentation
4.5.3 Categorical data
pandas provides a Categorical function and a category object type to Python. This type is analogous to the factor data type in R. It is meant to address categorical or discrete variables, where we need to use them in analyses. Categorical variables typically take on a small number of unique values, like gender, blood type, country of origin, race, etc.
You can create categorical Series in a couple of ways:
df = pd.DataFrame({
'A':3.,
'B':rng.random_sample(5),
'C': pd.Timestamp('20200512'),
'D': np.array([6] * 5),
'E': pd.Categorical(['yes','no','no','yes','no']),
'F': 'NIH'})
df['F'].astype('category')0 NIH
1 NIH
2 NIH
3 NIH
4 NIH
Name: F, dtype: category
Categories (1, object): [NIH]You can also create DataFrame’s where each column is categorical
df = pd.DataFrame({'A': list('abcd'), 'B': list('bdca')})
df_cat = df.astype('category')
df_cat.dtypesA category
B category
dtype: objectYou can explore categorical data in a variety of ways
count 4
unique 4
top d
freq 1
Name: A, dtype: objectd 1
b 1
a 1
c 1
Name: A, dtype: int64One issue with categories is that, if a particular level of a category is not seen before, it can create an error. So you can pre-specify the categories you expect
df_cat['B'] = pd.Categorical(list('aabb'), categories = ['a','b','c','d'])
df_cat['B'].value_counts()b 2
a 2
d 0
c 0
Name: B, dtype: int644.5.3.1 Re-organizing categories
In categorical data, there is often the concept of a “first” or “reference” category, and an ordering of categories. This tends to be important in both visualization as well as in regression modeling. Both aspects of a category can be addressed using the reorder_categories function.
In our earlier example, we can see that the A variable has 4 categories, with the “first” category being “a”.
0 a
1 b
2 c
3 d
Name: A, dtype: category
Categories (4, object): [a, b, c, d]Suppose we want to change this ordering to the reverse ordering, where “d” is the “first” category, and then it goes in reverse order.
0 a
1 b
2 c
3 d
Name: A, dtype: category
Categories (4, object): [d, c, b, a]4.5.4 Missing data
Both numpy and pandas allow for missing values, which are a reality in data science. The missing values are coded as np.nan. Let’s create some data and force some missing values
df = pd.DataFrame(np.random.randn(5, 3), index = ['a','c','e', 'f','g'], columns = ['one','two','three']) # pre-specify index and column names
df['four'] = 20 # add a column named "four", which will all be 20
df['five'] = df['one'] > 0
df one two three four five
a -0.706987 -0.821679 1.441257 20 False
c 1.297128 0.501395 0.572570 20 True
e -0.761507 1.469939 0.400255 20 False
f -0.910821 0.449404 0.588879 20 False
g -0.718350 -0.364237 1.793386 20 Falsedf2 = df.reindex(['a','b','c','d','e','f','g'])
df2.style.applymap(lambda x: 'background-color:yellow', subset = pd.IndexSlice[['b','d'],:])<pandas.io.formats.style.Styler object at 0x11cbd6040>The code above is creating new blank rows based on the new index values, some of which are present in the existing data and some of which are missing.
We can create masks of the data indicating where missing values reside in a data set.
one two three four five
a False False False False False
b True True True True True
c False False False False False
d True True True True True
e False False False False False
f False False False False False
g False False False False Falsea True
b False
c True
d False
e True
f True
g True
Name: one, dtype: boolWe can obtain complete data by dropping any row that has any missing value. This is called complete case analysis, and you should be very careful using it. It is only valid if we belive that the missingness is missing at random, and not related to some characteristic of the data or the data gathering process.
one two three four five
a -0.706987 -0.821679 1.441257 20.0 False
c 1.297128 0.501395 0.572570 20.0 True
e -0.761507 1.469939 0.400255 20.0 False
f -0.910821 0.449404 0.588879 20.0 False
g -0.718350 -0.364237 1.793386 20.0 FalseYou can also fill in, or impute, missing values. This can be done using a single value..
out1 = df2.fillna(value = 5)
out1.style.applymap(lambda x: 'background-color:yellow', subset = pd.IndexSlice[['b','d'],:])<pandas.io.formats.style.Styler object at 0x11cf5fca0>or a computed value like a column mean
df3 = df2.copy()
df3 = df3.select_dtypes(exclude=[object]) # remove non-numeric columns
out2 = df3.fillna(df3.mean()) # df3.mean() computes column-wise means
out2.style.applymap(lambda x: 'background-color:yellow', subset = pd.IndexSlice[['b','d'],:])<pandas.io.formats.style.Styler object at 0x11cf830d0>You can also impute based on the principle of last value carried forward which is common in time series. This means that the missing value is imputed with the previous recorded value.
out3 = df2.fillna(method = 'ffill') # Fill forward
out3.style.applymap(lambda x: 'background-color:yellow', subset = pd.IndexSlice[['b','d'],:])<pandas.io.formats.style.Styler object at 0x11cbeca60>out4 = df2.fillna(method = 'bfill') # Fill backward
out4.style.applymap(lambda x: 'background-color:yellow', subset = pd.IndexSlice[['b','d'],:])<pandas.io.formats.style.Styler object at 0x11cbd6ca0>4.6 Data transformation
4.6.1 Arithmetic operations
If you have a Series or DataFrame that is all numeric, you can add or multiply single numbers to all the elements together.
0 1 2 3 4
0 0.243690 1.076332 0.950349 1.373858 0.868005
1 2.712568 1.046159 -2.044472 -0.630508 0.945787
2 0.413522 1.938513 -0.195251 0.350383 1.381913
3 -0.927886 -1.882394 -2.163616 -0.142430 1.091522 0 1 2 3 4
0 6.243690 7.076332 6.950349 7.373858 6.868005
1 8.712568 7.046159 3.955528 5.369492 6.945787
2 6.413522 7.938513 5.804749 6.350383 7.381913
3 5.072114 4.117606 3.836384 5.857570 7.091522 0 1 2 3 4
0 -2.436896 -10.763319 -9.503492 -13.738582 -8.680051
1 -27.125677 -10.461590 20.444719 6.305081 -9.457871
2 -4.135220 -19.385126 1.952507 -3.503830 -13.819127
3 9.278864 18.823936 21.636155 1.424302 -10.915223If you have two compatible (same dimension) numeric DataFrames, you can add, subtract, multiply and divide elementwise
0 1 2 3 4
0 4.018707 5.687454 3.580436 6.782426 5.120555
1 5.253674 6.124992 1.169963 2.695430 5.307602
2 4.217585 5.948985 4.620141 4.040466 4.649786
3 3.890115 2.221573 1.245353 5.396740 5.127738 0 1 2 3 4
0 0.919932 4.963098 2.499501 7.430605 3.691235
1 6.892923 5.313267 -6.571821 -2.097031 4.125348
2 1.573064 7.774351 -0.940208 1.292943 4.515915
3 -4.470558 -7.725281 -7.375697 -0.788945 4.405619If you have a Series with the same number of elements as the number of columns of a DataFrame, you can do arithmetic operations, with each element of the Series acting upon each column of the DataFrame
0 1 2 3 4
0 1.243690 3.076332 3.950349 5.373858 5.868005
1 3.712568 3.046159 0.955528 3.369492 5.945787
2 1.413522 3.938513 2.804749 4.350383 6.381913
3 0.072114 0.117606 0.836384 3.857570 6.091522 0 1 2 3 4
0 0.243690 2.152664 2.851048 5.495433 4.340025
1 2.712568 2.092318 -6.133416 -2.522032 4.728935
2 0.413522 3.877025 -0.585752 1.401532 6.909563
3 -0.927886 -3.764787 -6.490847 -0.569721 5.457611This idea can be used to standardize a dataset, i.e. make each column have mean 0 and standard deviation 1.
0 1 2 3 4
0 -0.240828 0.318354 1.202702 1.326047 -0.899601
1 1.380224 0.300287 -0.783339 -1.013572 -0.556263
2 -0.129317 0.834602 0.442988 0.131384 1.368838
3 -1.010079 -1.453244 -0.862351 -0.443858 0.0870264.6.2 Concatenation of data sets
Let’s create some example data sets
df1 = pd.DataFrame({'A': ['a'+str(i) for i in range(4)],
'B': ['b'+str(i) for i in range(4)],
'C': ['c'+str(i) for i in range(4)],
'D': ['d'+str(i) for i in range(4)]})
df2 = pd.DataFrame({'A': ['a'+str(i) for i in range(4,8)],
'B': ['b'+str(i) for i in range(4,8)],
'C': ['c'+str(i) for i in range(4,8)],
'D': ['d'+str(i) for i in range(4,8)]})
df3 = pd.DataFrame({'A': ['a'+str(i) for i in range(8,12)],
'B': ['b'+str(i) for i in range(8,12)],
'C': ['c'+str(i) for i in range(8,12)],
'D': ['d'+str(i) for i in range(8,12)]})We can concatenate these DataFrame objects by row
A B C D
0 a0 b0 c0 d0
1 a1 b1 c1 d1
2 a2 b2 c2 d2
3 a3 b3 c3 d3
0 a4 b4 c4 d4
1 a5 b5 c5 d5
2 a6 b6 c6 d6
3 a7 b7 c7 d7
0 a8 b8 c8 d8
1 a9 b9 c9 d9
2 a10 b10 c10 d10
3 a11 b11 c11 d11This stacks the dataframes together. They are literally stacked, as is evidenced by the index values being repeated.
This same exercise can be done by the append function
A B C D
0 a0 b0 c0 d0
1 a1 b1 c1 d1
2 a2 b2 c2 d2
3 a3 b3 c3 d3
0 a4 b4 c4 d4
1 a5 b5 c5 d5
2 a6 b6 c6 d6
3 a7 b7 c7 d7
0 a8 b8 c8 d8
1 a9 b9 c9 d9
2 a10 b10 c10 d10
3 a11 b11 c11 d11Suppose we want to append a new row to df1. Lets create a new row.
A B C D 0
0 a0 b0 c0 d0 NaN
1 a1 b1 c1 d1 NaN
2 a2 b2 c2 d2 NaN
3 a3 b3 c3 d3 NaN
0 NaN NaN NaN NaN n1
1 NaN NaN NaN NaN n2
2 NaN NaN NaN NaN n3
3 NaN NaN NaN NaN n4That’s a lot of missing values. The issue is that the we don’t have column names in the new_row, and the indices are the same, so pandas tries to append it my making a new column. The solution is to make it a DataFrame.
A B C D
0 n1 n2 n3 n4 A B C D
0 a0 b0 c0 d0
1 a1 b1 c1 d1
2 a2 b2 c2 d2
3 a3 b3 c3 d3
0 n1 n2 n3 n4or
A B C D
0 a0 b0 c0 d0
1 a1 b1 c1 d1
2 a2 b2 c2 d2
3 a3 b3 c3 d3
0 n1 n2 n3 n44.6.2.1 Adding columns
A B C D A B C D A B C D
0 a0 b0 c0 d0 a4 b4 c4 d4 a8 b8 c8 d8
1 a1 b1 c1 d1 a5 b5 c5 d5 a9 b9 c9 d9
2 a2 b2 c2 d2 a6 b6 c6 d6 a10 b10 c10 d10
3 a3 b3 c3 d3 a7 b7 c7 d7 a11 b11 c11 d11The option axis=1 ensures that concatenation happens by columns. The default value axis = 0 concatenates by rows.
Let’s play a little game. Let’s change the column names of df2 and df3 so they are not the same as df1.
A B C D E F G H
0 a0 b0 c0 d0 NaN NaN NaN NaN
1 a1 b1 c1 d1 NaN NaN NaN NaN
2 a2 b2 c2 d2 NaN NaN NaN NaN
3 a3 b3 c3 d3 NaN NaN NaN NaN
0 NaN NaN NaN NaN a4 b4 c4 d4
1 NaN NaN NaN NaN a5 b5 c5 d5
2 NaN NaN NaN NaN a6 b6 c6 d6
3 NaN NaN NaN NaN a7 b7 c7 d7
0 a8 NaN NaN b8 NaN c8 NaN d8
1 a9 NaN NaN b9 NaN c9 NaN d9
2 a10 NaN NaN b10 NaN c10 NaN d10
3 a11 NaN NaN b11 NaN c11 NaN d11Now pandas ensures that all column names are represented in the new data frame, but with missing values where the row indices and column indices are mismatched. Some of this can be avoided by only joining on common columns. This is done using the join option ir concat. The default value is ’outer`, which is what you see. above
A D
0 a0 d0
1 a1 d1
2 a2 d2
3 a3 d3
0 a8 b8
1 a9 b9
2 a10 b10
3 a11 b11You can do the same thing when joining by rows, using axis = 0 and join="inner" to only join on rows with matching indices. Reminder that the indices are just labels and happen to be the row numbers by default.
4.6.3 Merging data sets
For this section we’ll use a set of data from a survey, also used by Daniel Chen in “Pandas for Everyone”
person = pd.read_csv('data/survey_person.csv')
site = pd.read_csv('data/survey_site.csv')
survey = pd.read_csv('data/survey_survey.csv')
visited = pd.read_csv('data/survey_visited.csv') ident personal family
0 dyer William Dyer
1 pb Frank Pabodie
2 lake Anderson Lake
3 roe Valentina Roerich
4 danforth Frank Danforth name lat long
0 DR-1 -49.85 -128.57
1 DR-3 -47.15 -126.72
2 MSK-4 -48.87 -123.40 taken person quant reading
0 619 dyer rad 9.82
1 619 dyer sal 0.13
2 622 dyer rad 7.80
3 622 dyer sal 0.09
4 734 pb rad 8.41
5 734 lake sal 0.05
6 734 pb temp -21.50
7 735 pb rad 7.22
8 735 NaN sal 0.06
9 735 NaN temp -26.00
10 751 pb rad 4.35
11 751 pb temp -18.50
12 751 lake sal 0.10
13 752 lake rad 2.19
14 752 lake sal 0.09
15 752 lake temp -16.00
16 752 roe sal 41.60
17 837 lake rad 1.46
18 837 lake sal 0.21
19 837 roe sal 22.50
20 844 roe rad 11.25 ident site dated
0 619 DR-1 1927-02-08
1 622 DR-1 1927-02-10
2 734 DR-3 1939-01-07
3 735 DR-3 1930-01-12
4 751 DR-3 1930-02-26
5 752 DR-3 NaN
6 837 MSK-4 1932-01-14
7 844 DR-1 1932-03-22There are basically four kinds of joins:
| pandas | R | SQL | Description |
|---|---|---|---|
| left | left_join | left outer | keep all rows on left |
| right | right_join | right outer | keep all rows on right |
| outer | outer_join | full outer | keep all rows from both |
| inner | inner_join | inner | keep only rows with common keys |
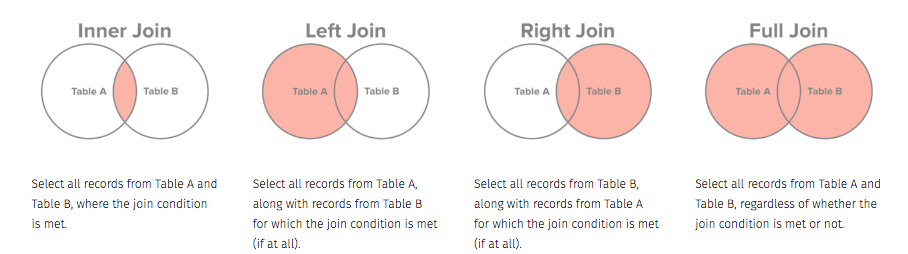
The terms left and right refer to which data set you call first and second respectively.
We start with an left join
taken person quant reading ident site dated
0 619 dyer rad 9.82 619 DR-1 1927-02-08
1 619 dyer sal 0.13 619 DR-1 1927-02-08
2 622 dyer rad 7.80 622 DR-1 1927-02-10
3 622 dyer sal 0.09 622 DR-1 1927-02-10
4 734 pb rad 8.41 734 DR-3 1939-01-07
5 734 lake sal 0.05 734 DR-3 1939-01-07
6 734 pb temp -21.50 734 DR-3 1939-01-07
7 735 pb rad 7.22 735 DR-3 1930-01-12
8 735 NaN sal 0.06 735 DR-3 1930-01-12
9 735 NaN temp -26.00 735 DR-3 1930-01-12
10 751 pb rad 4.35 751 DR-3 1930-02-26
11 751 pb temp -18.50 751 DR-3 1930-02-26
12 751 lake sal 0.10 751 DR-3 1930-02-26
13 752 lake rad 2.19 752 DR-3 NaN
14 752 lake sal 0.09 752 DR-3 NaN
15 752 lake temp -16.00 752 DR-3 NaN
16 752 roe sal 41.60 752 DR-3 NaN
17 837 lake rad 1.46 837 MSK-4 1932-01-14
18 837 lake sal 0.21 837 MSK-4 1932-01-14
19 837 roe sal 22.50 837 MSK-4 1932-01-14
20 844 roe rad 11.25 844 DR-1 1932-03-22Here, the left dataset is survey and the right one is visited. Since we’re doing a left join, we keed all the rows from survey and add columns from visited, matching on the common key, called “taken” in one dataset and “ident” in the other. Note that the rows of visited are repeated as needed to line up with all the rows with common “taken” values.
We can now add location information, where the common key is the site code
s2v2loc_merge = s2v_merge.merge(site, how = 'left', left_on = 'site', right_on = 'name')
print(s2v2loc_merge) taken person quant reading ident site dated name lat long
0 619 dyer rad 9.82 619 DR-1 1927-02-08 DR-1 -49.85 -128.57
1 619 dyer sal 0.13 619 DR-1 1927-02-08 DR-1 -49.85 -128.57
2 622 dyer rad 7.80 622 DR-1 1927-02-10 DR-1 -49.85 -128.57
3 622 dyer sal 0.09 622 DR-1 1927-02-10 DR-1 -49.85 -128.57
4 734 pb rad 8.41 734 DR-3 1939-01-07 DR-3 -47.15 -126.72
5 734 lake sal 0.05 734 DR-3 1939-01-07 DR-3 -47.15 -126.72
6 734 pb temp -21.50 734 DR-3 1939-01-07 DR-3 -47.15 -126.72
7 735 pb rad 7.22 735 DR-3 1930-01-12 DR-3 -47.15 -126.72
8 735 NaN sal 0.06 735 DR-3 1930-01-12 DR-3 -47.15 -126.72
9 735 NaN temp -26.00 735 DR-3 1930-01-12 DR-3 -47.15 -126.72
10 751 pb rad 4.35 751 DR-3 1930-02-26 DR-3 -47.15 -126.72
11 751 pb temp -18.50 751 DR-3 1930-02-26 DR-3 -47.15 -126.72
12 751 lake sal 0.10 751 DR-3 1930-02-26 DR-3 -47.15 -126.72
13 752 lake rad 2.19 752 DR-3 NaN DR-3 -47.15 -126.72
14 752 lake sal 0.09 752 DR-3 NaN DR-3 -47.15 -126.72
15 752 lake temp -16.00 752 DR-3 NaN DR-3 -47.15 -126.72
16 752 roe sal 41.60 752 DR-3 NaN DR-3 -47.15 -126.72
17 837 lake rad 1.46 837 MSK-4 1932-01-14 MSK-4 -48.87 -123.40
18 837 lake sal 0.21 837 MSK-4 1932-01-14 MSK-4 -48.87 -123.40
19 837 roe sal 22.50 837 MSK-4 1932-01-14 MSK-4 -48.87 -123.40
20 844 roe rad 11.25 844 DR-1 1932-03-22 DR-1 -49.85 -128.57Lastly, we add the person information to this dataset.
merged = s2v2loc_merge.merge(person, how = 'left', left_on = 'person', right_on = 'ident')
print(merged.head()) taken person quant reading ... long ident_y personal family
0 619 dyer rad 9.82 ... -128.57 dyer William Dyer
1 619 dyer sal 0.13 ... -128.57 dyer William Dyer
2 622 dyer rad 7.80 ... -128.57 dyer William Dyer
3 622 dyer sal 0.09 ... -128.57 dyer William Dyer
4 734 pb rad 8.41 ... -126.72 pb Frank Pabodie
[5 rows x 13 columns]You can merge based on multiple columns as long as they match up.
ps = person.merge(survey, left_on = 'ident', right_on = 'person')
vs = visited.merge(survey, left_on = 'ident', right_on = 'taken')
print(ps) ident personal family taken person quant reading
0 dyer William Dyer 619 dyer rad 9.82
1 dyer William Dyer 619 dyer sal 0.13
2 dyer William Dyer 622 dyer rad 7.80
3 dyer William Dyer 622 dyer sal 0.09
4 pb Frank Pabodie 734 pb rad 8.41
5 pb Frank Pabodie 734 pb temp -21.50
6 pb Frank Pabodie 735 pb rad 7.22
7 pb Frank Pabodie 751 pb rad 4.35
8 pb Frank Pabodie 751 pb temp -18.50
9 lake Anderson Lake 734 lake sal 0.05
10 lake Anderson Lake 751 lake sal 0.10
11 lake Anderson Lake 752 lake rad 2.19
12 lake Anderson Lake 752 lake sal 0.09
13 lake Anderson Lake 752 lake temp -16.00
14 lake Anderson Lake 837 lake rad 1.46
15 lake Anderson Lake 837 lake sal 0.21
16 roe Valentina Roerich 752 roe sal 41.60
17 roe Valentina Roerich 837 roe sal 22.50
18 roe Valentina Roerich 844 roe rad 11.25 ident site dated taken person quant reading
0 619 DR-1 1927-02-08 619 dyer rad 9.82
1 619 DR-1 1927-02-08 619 dyer sal 0.13
2 622 DR-1 1927-02-10 622 dyer rad 7.80
3 622 DR-1 1927-02-10 622 dyer sal 0.09
4 734 DR-3 1939-01-07 734 pb rad 8.41
5 734 DR-3 1939-01-07 734 lake sal 0.05
6 734 DR-3 1939-01-07 734 pb temp -21.50
7 735 DR-3 1930-01-12 735 pb rad 7.22
8 735 DR-3 1930-01-12 735 NaN sal 0.06
9 735 DR-3 1930-01-12 735 NaN temp -26.00
10 751 DR-3 1930-02-26 751 pb rad 4.35
11 751 DR-3 1930-02-26 751 pb temp -18.50
12 751 DR-3 1930-02-26 751 lake sal 0.10
13 752 DR-3 NaN 752 lake rad 2.19
14 752 DR-3 NaN 752 lake sal 0.09
15 752 DR-3 NaN 752 lake temp -16.00
16 752 DR-3 NaN 752 roe sal 41.60
17 837 MSK-4 1932-01-14 837 lake rad 1.46
18 837 MSK-4 1932-01-14 837 lake sal 0.21
19 837 MSK-4 1932-01-14 837 roe sal 22.50
20 844 DR-1 1932-03-22 844 roe rad 11.25ps_vs = ps.merge(vs,
left_on = ['ident','taken', 'quant','reading'],
right_on = ['person','ident','quant','reading']) # The keys need to correspond
ps_vs.head() ident_x personal family taken_x ... site dated taken_y person_y
0 dyer William Dyer 619 ... DR-1 1927-02-08 619 dyer
1 dyer William Dyer 619 ... DR-1 1927-02-08 619 dyer
2 dyer William Dyer 622 ... DR-1 1927-02-10 622 dyer
3 dyer William Dyer 622 ... DR-1 1927-02-10 622 dyer
4 pb Frank Pabodie 734 ... DR-3 1939-01-07 734 pb
[5 rows x 12 columns]Note that since there are common column names, the merge appends _x and _y to denote which column came from the left and right, respectively.
4.6.4 Tidy data principles and reshaping datasets
The tidy data principle is a principle espoused by Dr. Hadley Wickham, one of the foremost R developers. Tidy data is a structure for datasets to make them more easily analyzed on computers. The basic principles are
- Each row is an observation
- Each column is a variable
- Each type of observational unit forms a table
Tidy data is tidy in one way. Untidy data can be untidy in many ways
Let’s look at some examples.
from glob import glob
filenames = sorted(glob('data/table*.csv')) # find files matching pattern. I know there are 6 of them
table1, table2, table3, table4a, table4b, table5 = [pd.read_csv(f) for f in filenames] # Use a list comprehensionThis code imports data from 6 files matching a pattern. Python allows multiple assignments on the left of the =, and as each dataset is imported, it gets assigned in order to the variables on the left. In the second line I sort the file names so that they match the order in which I’m storing them in the 3rd line. The function glob does pattern-matching of file names.
The following tables refer to the number of TB cases and population in Afghanistan, Brazil and China in 1999 and 2000
country year cases population
0 Afghanistan 1999 745 19987071
1 Afghanistan 2000 2666 20595360
2 Brazil 1999 37737 172006362
3 Brazil 2000 80488 174504898
4 China 1999 212258 1272915272
5 China 2000 213766 1280428583 country year type count
0 Afghanistan 1999 cases 745
1 Afghanistan 1999 population 19987071
2 Afghanistan 2000 cases 2666
3 Afghanistan 2000 population 20595360
4 Brazil 1999 cases 37737
5 Brazil 1999 population 172006362
6 Brazil 2000 cases 80488
7 Brazil 2000 population 174504898
8 China 1999 cases 212258
9 China 1999 population 1272915272
10 China 2000 cases 213766
11 China 2000 population 1280428583 country year rate
0 Afghanistan 1999 745/19987071
1 Afghanistan 2000 2666/20595360
2 Brazil 1999 37737/172006362
3 Brazil 2000 80488/174504898
4 China 1999 212258/1272915272
5 China 2000 213766/1280428583 country 1999 2000
0 Afghanistan 745 2666
1 Brazil 37737 80488
2 China 212258 213766 country 1999 2000
0 Afghanistan 19987071 20595360
1 Brazil 172006362 174504898
2 China 1272915272 1280428583 country century year rate
0 Afghanistan 19 99 745/19987071
1 Afghanistan 20 0 2666/20595360
2 Brazil 19 99 37737/172006362
3 Brazil 20 0 80488/174504898
4 China 19 99 212258/1272915272
5 China 20 0 213766/1280428583Exercise: Describe why and why not each of these datasets are tidy.
4.6.5 Melting (unpivoting) data
Melting is the operation of collapsing multiple columns into 2 columns, where one column is formed by the old column names, and the other by the corresponding values. Some columns may be kept fixed and their data are repeated to maintain the interrelationships between the variables.
We’ll start with loading some data on income and religion in the US from the Pew Research Center.
religion <$10k $10-20k ... $100-150k >150k Don't know/refused
0 Agnostic 27 34 ... 109 84 96
1 Atheist 12 27 ... 59 74 76
2 Buddhist 27 21 ... 39 53 54
3 Catholic 418 617 ... 792 633 1489
4 Don’t know/refused 15 14 ... 17 18 116
[5 rows x 11 columns]This dataset is considered in “wide” format. There are several issues with it, including the fact that column headers have data. Those column headers are income groups, that should be a column by tidy principles. Our job is to turn this dataset into “long” format with a column for income group.
We will use the function melt to achieve this. This takes a few parameters:
- id_vars is a list of variables that will remain as is
- value_vars is a list of column nmaes that we will melt (or unpivot). By default, it will melt all columns not mentioned in id_vars
- var_name is a string giving the name of the new column created by the headers (default:
variable) - value_name is a string giving the name of the new column created by the values (default:
value)
pew_long = pew.melt(id_vars = ['religion'], var_name = 'income_group', value_name = 'count')
print(pew_long.head()) religion income_group count
0 Agnostic <$10k 27
1 Atheist <$10k 12
2 Buddhist <$10k 27
3 Catholic <$10k 418
4 Don’t know/refused <$10k 154.6.6 Separating columns containing multiple variables
We will use an Ebola dataset to illustrate this principle
Date Day ... Deaths_Spain Deaths_Mali
0 1/5/2015 289 ... NaN NaN
1 1/4/2015 288 ... NaN NaN
2 1/3/2015 287 ... NaN NaN
3 1/2/2015 286 ... NaN NaN
4 12/31/2014 284 ... NaN NaN
[5 rows x 18 columns]Note that for each country we have two columns – one for cases (number infected) and one for deaths. Ideally we want one column for country, one for cases and one for deaths.
The first step will be to melt this data sets so that the column headers in question from a column and the corresponding data forms a second column.
Date Day variable value
0 1/5/2015 289 Cases_Guinea 2776.0
1 1/4/2015 288 Cases_Guinea 2775.0
2 1/3/2015 287 Cases_Guinea 2769.0
3 1/2/2015 286 Cases_Guinea NaN
4 12/31/2014 284 Cases_Guinea 2730.0We now need to split the data in the variable column to make two columns. One will contain the country name and the other either Cases or Deaths. We will use some string manipulation functions that we will see later to achieve this.
variable_split = ebola_long['variable'].str.split('_', expand=True) # split on the `_` character
print(variable_split[:5]) 0 1
0 Cases Guinea
1 Cases Guinea
2 Cases Guinea
3 Cases Guinea
4 Cases GuineaThe expand=True option forces the creation of an DataFrame rather than a list
<class 'pandas.core.frame.DataFrame'>We can now concatenate this to the original data
variable_split.columns = ['status','country']
ebola_parsed = pd.concat([ebola_long, variable_split], axis = 1)
ebola_parsed.drop('variable', axis = 1, inplace=True) # Remove the column named "variable" and replace the old data with the new one in the same location
print(ebola_parsed.head()) Date Day value status country
0 1/5/2015 289 2776.0 Cases Guinea
1 1/4/2015 288 2775.0 Cases Guinea
2 1/3/2015 287 2769.0 Cases Guinea
3 1/2/2015 286 NaN Cases Guinea
4 12/31/2014 284 2730.0 Cases Guinea4.6.7 Pivot/spread datasets
If we wanted to, we could also make two columns based on cases and deaths, so for each country and date you could easily read off the cases and deaths. This is achieved using the pivot_table function.
In the pivot_table syntax, index refers to the columns we don’t want to change, columns refers to the column whose values will form the column names of the new columns, and values is the name of the column that will form the values in the pivoted dataset.
status Cases Deaths
Date Day country
1/2/2015 286 Liberia 8157.0 3496.0
1/3/2015 287 Guinea 2769.0 1767.0
Liberia 8166.0 3496.0
SierraLeone 9722.0 2915.0
1/4/2015 288 Guinea 2775.0 1781.0
... ... ...
9/7/2014 169 Liberia 2081.0 1137.0
Nigeria 21.0 8.0
Senegal 3.0 0.0
SierraLeone 1424.0 524.0
9/9/2014 171 Liberia 2407.0 NaN
[375 rows x 2 columns]This creates something called MultiIndex in the pandas DataFrame. This is useful in some advanced cases, but here, we just want a normal DataFrame back. We can achieve that by using the reset_index function.
ebola_parsed.pivot_table(index = ['Date','Day','country'], columns = 'status', values = 'value').reset_index()status Date Day country Cases Deaths
0 1/2/2015 286 Liberia 8157.0 3496.0
1 1/3/2015 287 Guinea 2769.0 1767.0
2 1/3/2015 287 Liberia 8166.0 3496.0
3 1/3/2015 287 SierraLeone 9722.0 2915.0
4 1/4/2015 288 Guinea 2775.0 1781.0
.. ... ... ... ... ...
370 9/7/2014 169 Liberia 2081.0 1137.0
371 9/7/2014 169 Nigeria 21.0 8.0
372 9/7/2014 169 Senegal 3.0 0.0
373 9/7/2014 169 SierraLeone 1424.0 524.0
374 9/9/2014 171 Liberia 2407.0 NaN
[375 rows x 5 columns]Pivoting is a 2-column to many-column operation, with the number of columns formed depending on the number of unique values present in the column of the original data that is entered into the columns argument of pivot_table
Exercise: Load the file weather.csv into Python and work on making it a tidy dataset. It requires melting and pivoting. The dataset comprises of the maximun and minimum temperatures recorded each day in 2010. There are lots of missing value. Ultimately we want columns for days of the month, maximum temperature and minimum tempearture along with the location ID, the year and the month.
4.7 Data aggregation and split-apply-combine
We’ll use the Gapminder dataset for this section
df = pd.read_csv('data/gapminder.tsv', sep = '\t') # data is tab-separated, so we use `\t` to specify thatThe paradigm we will be exploring is often called split-apply-combine or MapReduce or grouped aggregation. The basic idea is that you split a data set up by some feature, apply a recipe to each piece, compute the result, and then put the results back together into a dataset. This can be described in teh following schematic.
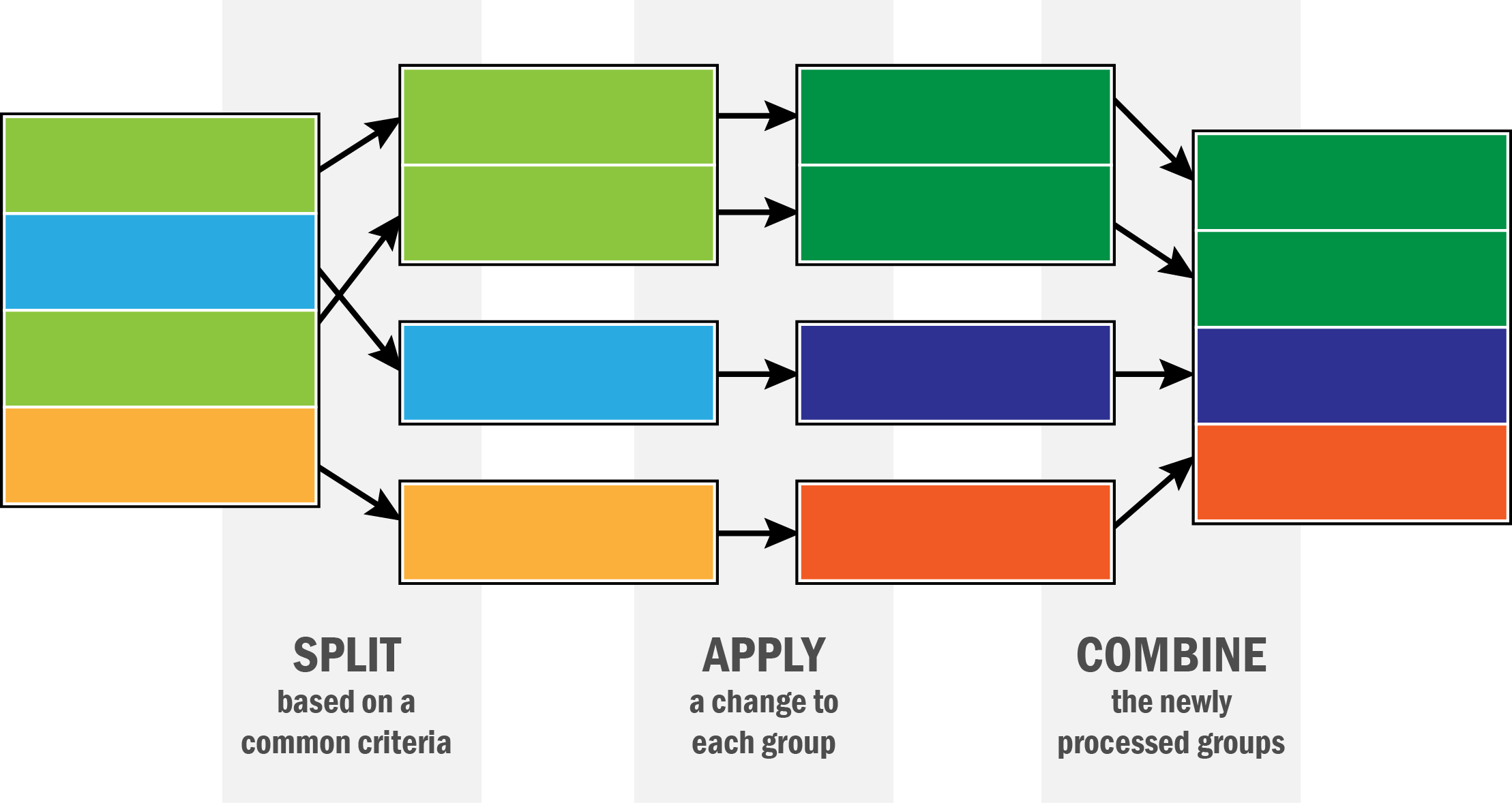
pandas is set up for this. It features the groupby function that allows the “split” part of the operation. We can then apply a function to each part and put it back together. Let’s see how.
country continent year lifeExp pop gdpPercap
0 Afghanistan Asia 1952 28.801 8425333 779.445314
1 Afghanistan Asia 1957 30.332 9240934 820.853030
2 Afghanistan Asia 1962 31.997 10267083 853.100710
3 Afghanistan Asia 1967 34.020 11537966 836.197138
4 Afghanistan Asia 1972 36.088 13079460 739.981106'This dataset has 142 countries in it'One of the variables in this dataset is life expectancy at birth, lifeExp. Suppose we want to find the average life expectancy of each country over the period of study.
country
Afghanistan 37.478833
Albania 68.432917
Algeria 59.030167
Angola 37.883500
Argentina 69.060417
...
Vietnam 57.479500
West Bank and Gaza 60.328667
Yemen, Rep. 46.780417
Zambia 45.996333
Zimbabwe 52.663167
Name: lifeExp, Length: 142, dtype: float64So what’s going on here? First, we use the groupby function, telling pandas to split the dataset up by values of the column country.
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x11cfb1b50>pandas won’t show you the actual data, but will tell you that it is a grouped dataframe object. This means that each element of this object is a DataFrame with data from one country.
142 country continent year lifeExp pop gdpPercap
1596 United Kingdom Europe 1952 69.180 50430000 9979.508487
1597 United Kingdom Europe 1957 70.420 51430000 11283.177950
1598 United Kingdom Europe 1962 70.760 53292000 12477.177070
1599 United Kingdom Europe 1967 71.360 54959000 14142.850890
1600 United Kingdom Europe 1972 72.010 56079000 15895.116410
1601 United Kingdom Europe 1977 72.760 56179000 17428.748460
1602 United Kingdom Europe 1982 74.040 56339704 18232.424520
1603 United Kingdom Europe 1987 75.007 56981620 21664.787670
1604 United Kingdom Europe 1992 76.420 57866349 22705.092540
1605 United Kingdom Europe 1997 77.218 58808266 26074.531360
1606 United Kingdom Europe 2002 78.471 59912431 29478.999190
1607 United Kingdom Europe 2007 79.425 60776238 33203.261280<class 'pandas.core.frame.DataFrame'>73.9225833333333273.92258333333332Let’s look at if life expectancy has gone up over time, by continent
continent year
Africa 1952 39.135500
1957 41.266346
1962 43.319442
1967 45.334538
1972 47.450942
1977 49.580423
1982 51.592865
1987 53.344788
1992 53.629577
1997 53.598269
2002 53.325231
2007 54.806038
Americas 1952 53.279840
1957 55.960280
1962 58.398760
1967 60.410920
1972 62.394920
1977 64.391560
1982 66.228840
1987 68.090720
1992 69.568360
1997 71.150480
2002 72.422040
2007 73.608120
Asia 1952 46.314394
1957 49.318544
1962 51.563223
1967 54.663640
1972 57.319269
1977 59.610556
1982 62.617939
1987 64.851182
1992 66.537212
1997 68.020515
2002 69.233879
2007 70.728485
Europe 1952 64.408500
1957 66.703067
1962 68.539233
1967 69.737600
1972 70.775033
1977 71.937767
1982 72.806400
1987 73.642167
1992 74.440100
1997 75.505167
2002 76.700600
2007 77.648600
Oceania 1952 69.255000
1957 70.295000
1962 71.085000
1967 71.310000
1972 71.910000
1977 72.855000
1982 74.290000
1987 75.320000
1992 76.945000
1997 78.190000
2002 79.740000
2007 80.719500
Name: lifeExp, dtype: float64avg_lifeexp_continent_yr = df.groupby(['continent','year']).lifeExp.mean().reset_index()
avg_lifeexp_continent_yr continent year lifeExp
0 Africa 1952 39.135500
1 Africa 1957 41.266346
2 Africa 1962 43.319442
3 Africa 1967 45.334538
4 Africa 1972 47.450942
5 Africa 1977 49.580423
6 Africa 1982 51.592865
7 Africa 1987 53.344788
8 Africa 1992 53.629577
9 Africa 1997 53.598269
10 Africa 2002 53.325231
11 Africa 2007 54.806038
12 Americas 1952 53.279840
13 Americas 1957 55.960280
14 Americas 1962 58.398760
15 Americas 1967 60.410920
16 Americas 1972 62.394920
17 Americas 1977 64.391560
18 Americas 1982 66.228840
19 Americas 1987 68.090720
20 Americas 1992 69.568360
21 Americas 1997 71.150480
22 Americas 2002 72.422040
23 Americas 2007 73.608120
24 Asia 1952 46.314394
25 Asia 1957 49.318544
26 Asia 1962 51.563223
27 Asia 1967 54.663640
28 Asia 1972 57.319269
29 Asia 1977 59.610556
30 Asia 1982 62.617939
31 Asia 1987 64.851182
32 Asia 1992 66.537212
33 Asia 1997 68.020515
34 Asia 2002 69.233879
35 Asia 2007 70.728485
36 Europe 1952 64.408500
37 Europe 1957 66.703067
38 Europe 1962 68.539233
39 Europe 1967 69.737600
40 Europe 1972 70.775033
41 Europe 1977 71.937767
42 Europe 1982 72.806400
43 Europe 1987 73.642167
44 Europe 1992 74.440100
45 Europe 1997 75.505167
46 Europe 2002 76.700600
47 Europe 2007 77.648600
48 Oceania 1952 69.255000
49 Oceania 1957 70.295000
50 Oceania 1962 71.085000
51 Oceania 1967 71.310000
52 Oceania 1972 71.910000
53 Oceania 1977 72.855000
54 Oceania 1982 74.290000
55 Oceania 1987 75.320000
56 Oceania 1992 76.945000
57 Oceania 1997 78.190000
58 Oceania 2002 79.740000
59 Oceania 2007 80.719500<class 'pandas.core.frame.DataFrame'>The aggregation function, in this case mean, does both the “apply” and “combine” parts of the process.
We can do quick aggregations with pandas
count mean std ... 50% 75% max
continent ...
Africa 624.0 48.865330 9.150210 ... 47.7920 54.41150 76.442
Americas 300.0 64.658737 9.345088 ... 67.0480 71.69950 80.653
Asia 396.0 60.064903 11.864532 ... 61.7915 69.50525 82.603
Europe 360.0 71.903686 5.433178 ... 72.2410 75.45050 81.757
Oceania 24.0 74.326208 3.795611 ... 73.6650 77.55250 81.235
[5 rows x 8 columns] country year lifeExp pop gdpPercap
continent
Africa Algeria 2002 70.994 31287142 5288.040382
Americas Argentina 2002 74.340 38331121 8797.640716
Asia Afghanistan 2002 42.129 25268405 726.734055
Europe Albania 2002 75.651 3508512 4604.211737
Oceania Australia 2002 80.370 19546792 30687.754730You can also use functions from other modules, or your own functions in this aggregation work.
continent
Africa 48.865330
Americas 64.658737
Asia 60.064903
Europe 71.903686
Oceania 74.326208
Name: lifeExp, dtype: float64def my_mean(values):
n = len(values)
sum = 0
for value in values:
sum += value
return(sum/n)
df.groupby('continent').lifeExp.agg(my_mean)continent
Africa 48.865330
Americas 64.658737
Asia 60.064903
Europe 71.903686
Oceania 74.326208
Name: lifeExp, dtype: float64You can do many functions at once
count_nonzero mean std
year
1952 142.0 49.057620 12.225956
1957 142.0 51.507401 12.231286
1962 142.0 53.609249 12.097245
1967 142.0 55.678290 11.718858
1972 142.0 57.647386 11.381953
1977 142.0 59.570157 11.227229
1982 142.0 61.533197 10.770618
1987 142.0 63.212613 10.556285
1992 142.0 64.160338 11.227380
1997 142.0 65.014676 11.559439
2002 142.0 65.694923 12.279823
2007 142.0 67.007423 12.073021You can also aggregate on different columns at the same time by passing a dict to the agg function
year lifeExp pop gdpPercap
0 1952 49.057620 3943953.0 1968.528344
1 1957 51.507401 4282942.0 2173.220291
2 1962 53.609249 4686039.5 2335.439533
3 1967 55.678290 5170175.5 2678.334741
4 1972 57.647386 5877996.5 3339.129407
5 1977 59.570157 6404036.5 3798.609244
6 1982 61.533197 7007320.0 4216.228428
7 1987 63.212613 7774861.5 4280.300366
8 1992 64.160338 8688686.5 4386.085502
9 1997 65.014676 9735063.5 4781.825478
10 2002 65.694923 10372918.5 5319.804524
11 2007 67.007423 10517531.0 6124.3711094.7.0.1 Transformation
You can do grouped transformations using this same method. We will compute the z-score for each year, i.e. we will substract the average life expectancy and divide by the standard deviation
0 -1.662719
1 -1.737377
2 -1.792867
3 -1.854699
4 -1.900878
...
1699 -0.081910
1700 -0.338167
1701 -1.580537
1702 -2.100756
1703 -1.955077
Name: lifeExp, Length: 1704, dtype: float64year
1952 -5.165078e-16
1957 2.902608e-17
1962 2.404180e-16
1967 -6.108181e-16
1972 1.784566e-16
1977 -9.456442e-16
1982 -1.623310e-16
1987 6.687725e-16
1992 5.457293e-16
1997 8.787963e-16
2002 5.254013e-16
2007 4.925637e-16
Name: lifeExp_z, dtype: float644.7.0.2 Filter
We can split the dataset by values of one variable, and filter out those splits that fail some criterion. The following code only keeps countries with a population of at least 10 million at some point during the study period
country continent year lifeExp pop gdpPercap lifeExp_z
0 Afghanistan Asia 1952 28.801 8425333 779.445314 -1.662719
1 Afghanistan Asia 1957 30.332 9240934 820.853030 -1.737377
2 Afghanistan Asia 1962 31.997 10267083 853.100710 -1.792867
3 Afghanistan Asia 1967 34.020 11537966 836.197138 -1.854699
4 Afghanistan Asia 1972 36.088 13079460 739.981106 -1.900878
... ... ... ... ... ... ... ...
1699 Zimbabwe Africa 1987 62.351 9216418 706.157306 -0.081910
1700 Zimbabwe Africa 1992 60.377 10704340 693.420786 -0.338167
1701 Zimbabwe Africa 1997 46.809 11404948 792.449960 -1.580537
1702 Zimbabwe Africa 2002 39.989 11926563 672.038623 -2.100756
1703 Zimbabwe Africa 2007 43.487 12311143 469.709298 -1.955077
[924 rows x 7 columns]