Lecture 4: Data Visualization
BIOF 339
October 3, 2018
Data Visualization in R
Gallery
Gallery
Gallery
Gallery
Manhattan plot
Gallery
Circular Manhattan plot
Gallery
Maps
Gallery
Interactive graphs
Gallery
Interactive graphs
# Date[1:41], format: "2018-08-30" "2018-08-31" "2018-09-01" "2018-09-02" "2018-09-03" ...Gallery
Animation
ggplot2
We’re making the decision to use ggplot2 for graphics
- Makes pretty good formatting choices out of the box
- Works like pipes!!
- Is declarative (tell it what you want) without getting caught up in minutae
- Strongly leverages data frames (good practice)
- Fast enough
- There are good templates if you want to change the look
Introduction to ggplot2
# install.packages('tidyverse')
library(ggplot2)Introduction to ggplot2
The ggplot2 package is a very flexible and (to me) intuitive way of visualizing data. It is based on the concept of layering elements on a canvas.
This idea of layering graphics on a canvas is, to me, a nice way of building graphs
Introduction to ggplot2
You need:
- A
data.frameobject - Aesthetic mappings (aes) to say what data is used for what purpose in the viz
- x- and y-direction
- shapes, colors, lines
- A geometry object (geom) to say what to draw
- You can “layer” geoms on each other to build plots
Introduction to ggplot2
ggplot used pipes before pipes were a thing.
However, it uses the + symbol for piping rather than the %>% operator, since it pre-dates the tidyverse
Introduction to ggplot2
library(ggplot2)
ggplot(mtcars, aes(x = wt, y = mpg)) + geom_point()- A
data.frameobject: mtcars - Aesthetic mapping:
- x-axis: wt
- y-axis: mpg
- Geometry:
- geom_point: draw points
Introduction to ggplot2
library(ggplot2)
ggplot(mtcars, aes(x = wt, y = mpg)) + geom_point()+ geom_smooth()- A
data.frameobject: mtcars - Aesthetic mapping:
- x-axis: wt
- y-axis: mpg
- Geometry:
- geom_point: draw points
- geom_smooth: Add a layer which draws a best-fitting line
ggplot2 examples
We will use the two data sets:
data_spine <- read.csv('http://www.araastat.com/BIOF339_PracticalR/
Lectures/lecturedataframe_data/Dataset_spine.csv',
stringsAsFactors = F)
data_brca <- read.csv('http://www.araastat.com/BIOF339_PracticalR/
Lectures/lecturedataframe_data/
clinical_data_breast_cancer_modified.csv',
stringsAsFactors = F)Plotting one variable
Histograms
ggplot(data_brca, aes(x = Age.at.Initial.Pathologic.Diagnosis)) +
geom_histogram() 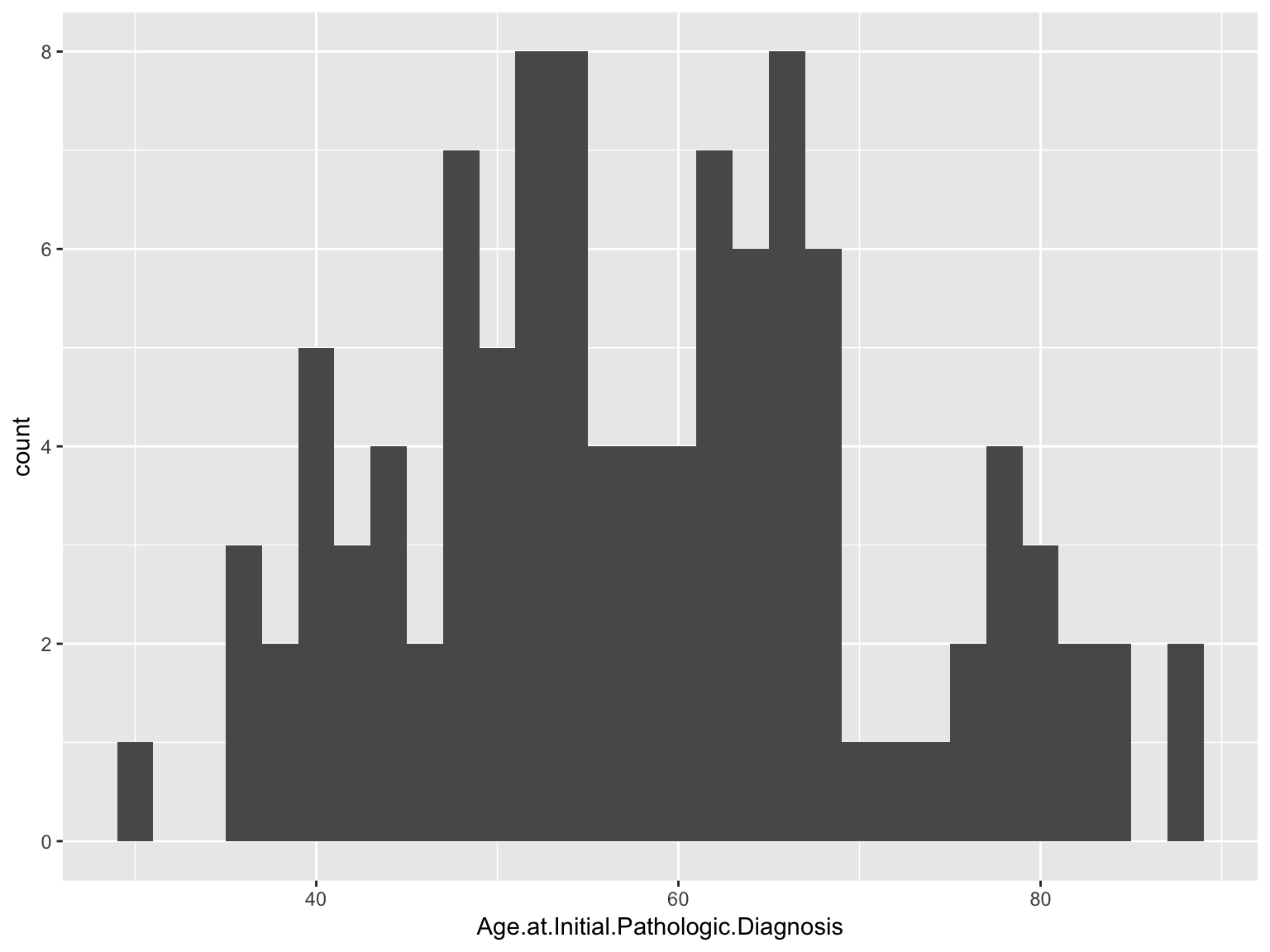
Histograms
ggplot(data_brca, aes(x = Age.at.Initial.Pathologic.Diagnosis)) +
geom_histogram(binwidth=4)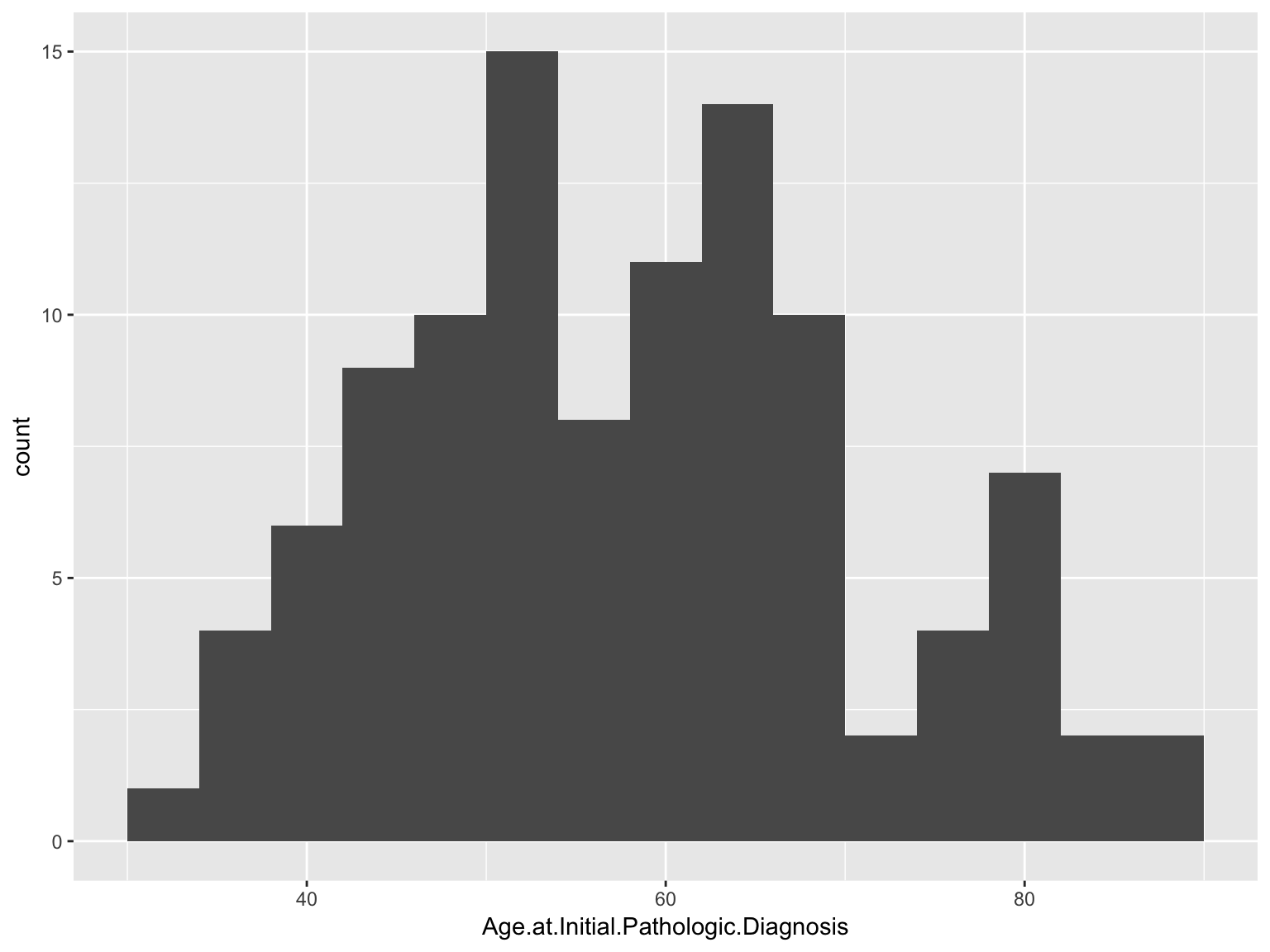
Density plot
ggplot(data_brca, aes(x = Age.at.Initial.Pathologic.Diagnosis)) +
geom_density()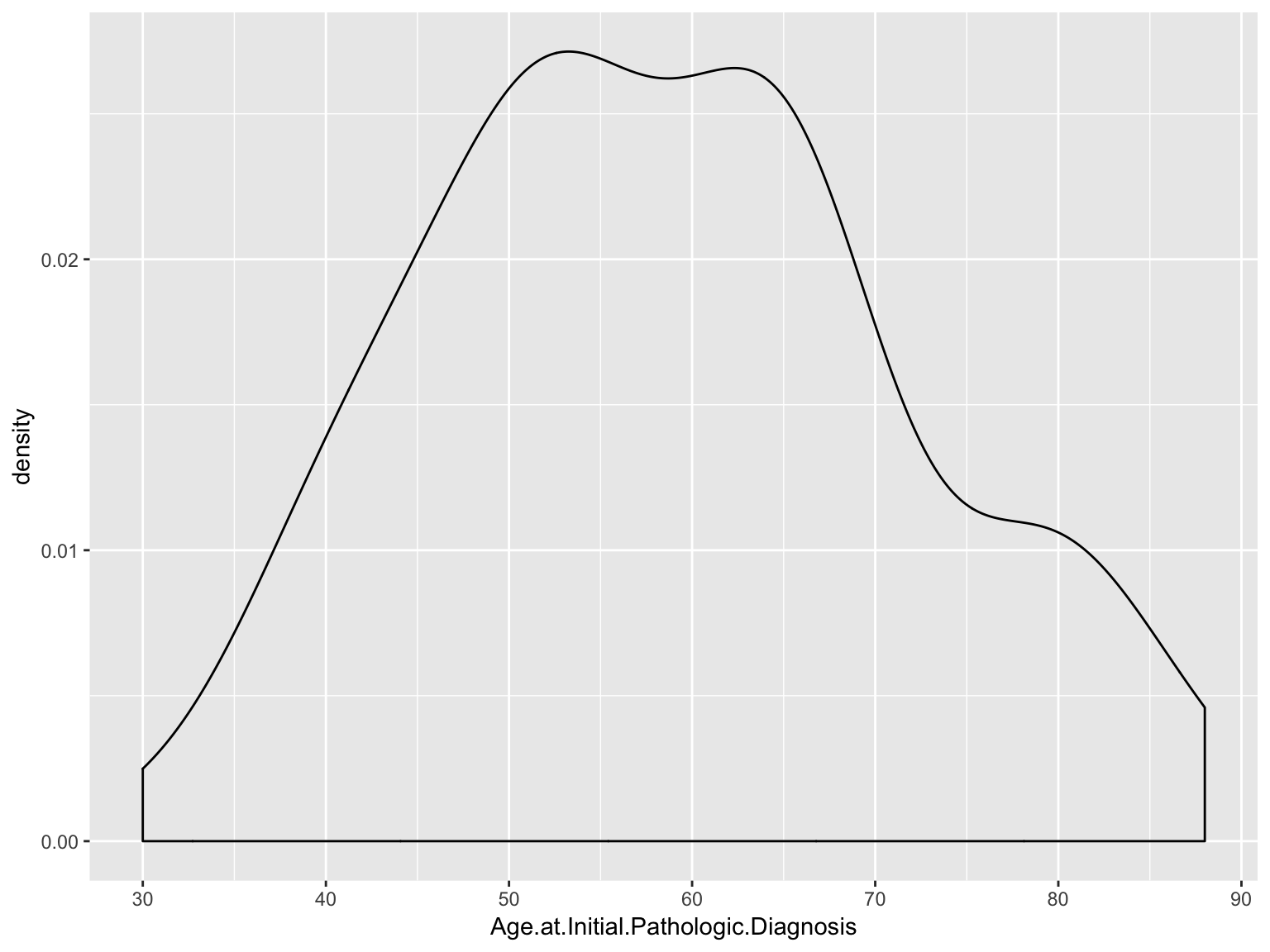
Bar plot
ggplot(data_brca, aes(x = Tumor))+geom_bar()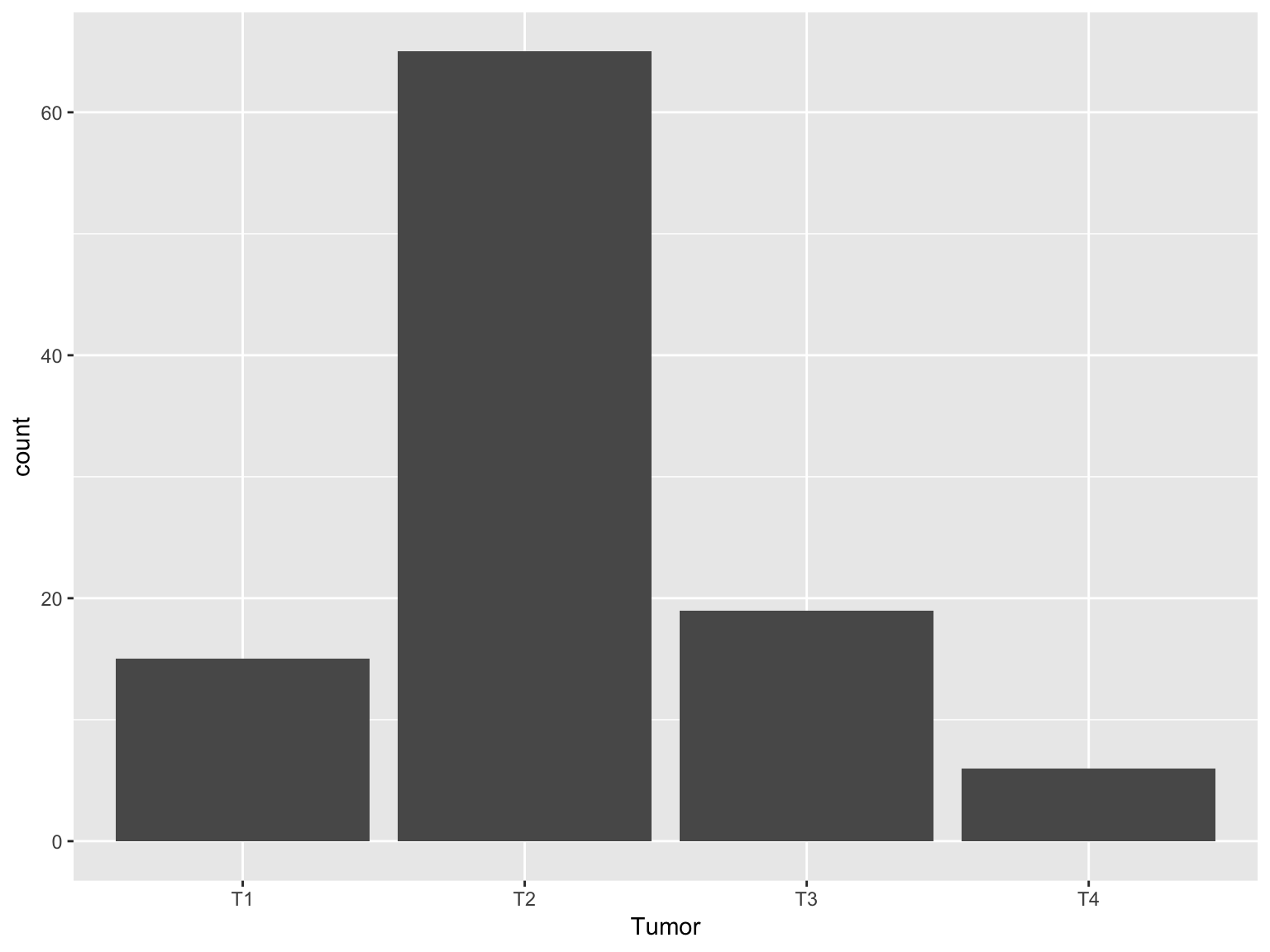
Exercise
Exercise
Using the mtcars dataset in R, create:
- A histogram of the fuel efficiences (
mpg) in the data set - A bar plot of frequencies of number of cylinders (
cyl) in the car
Solution
ggplot(mtcars, aes(x = mpg)) + geom_histogram(binwidth=3)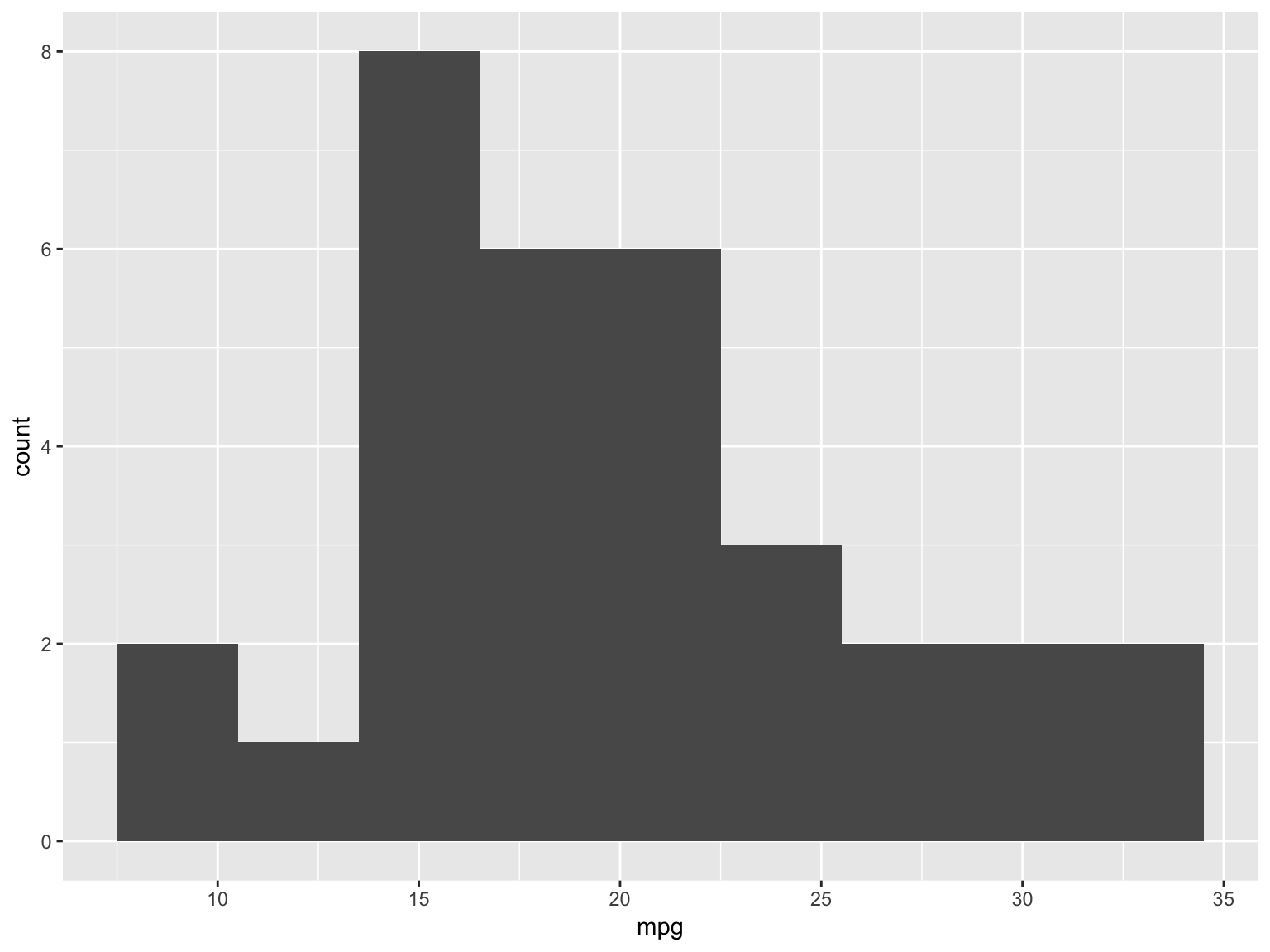
# ggplot(mtcars) + geom_histogram(aes(x = mpg), binwidth = 3)Solution
ggplot(mtcars, aes(x = factor(cyl))) + geom_bar()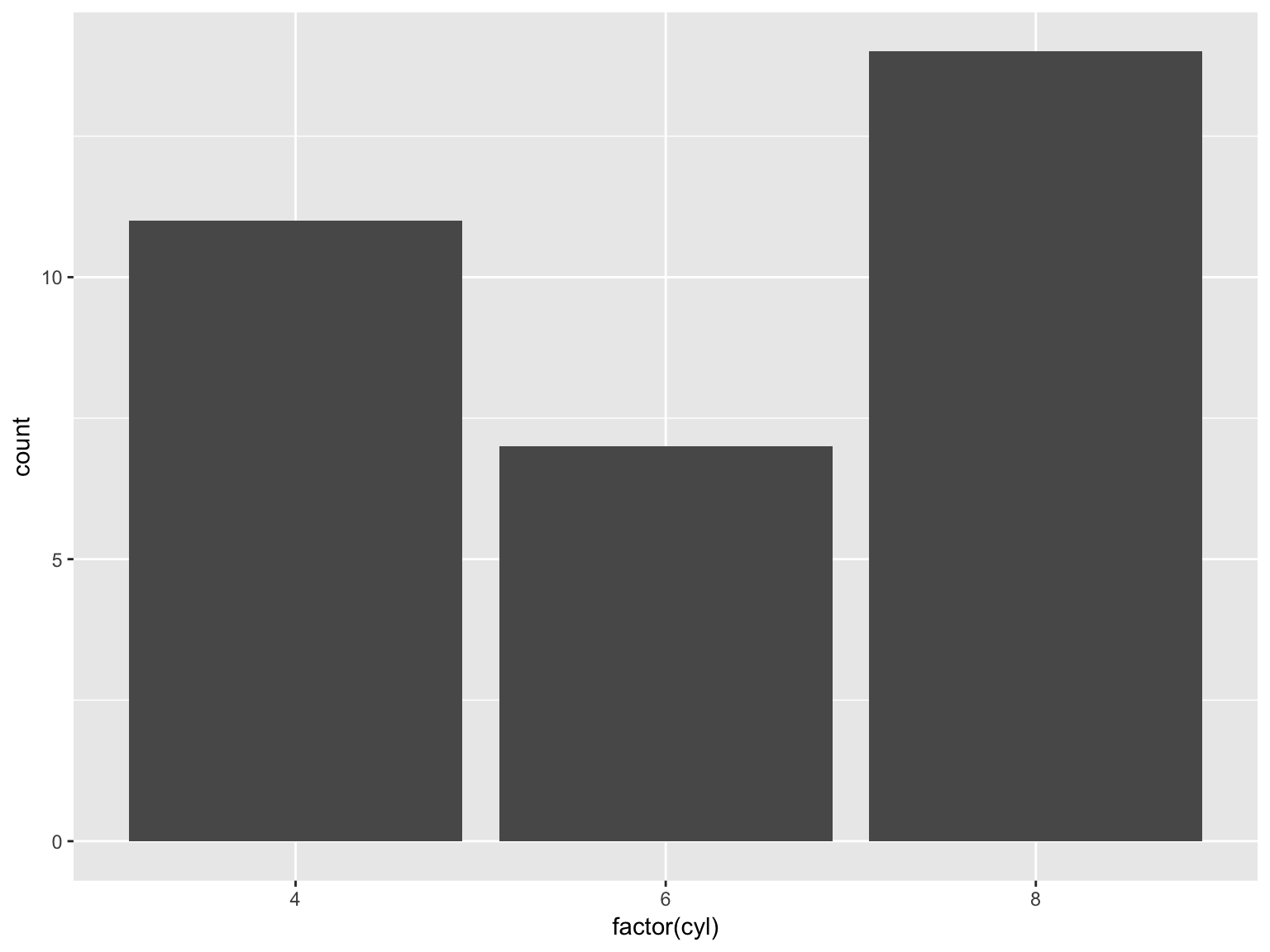
Two continuous variables
Scatter plots
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope)) +
geom_point()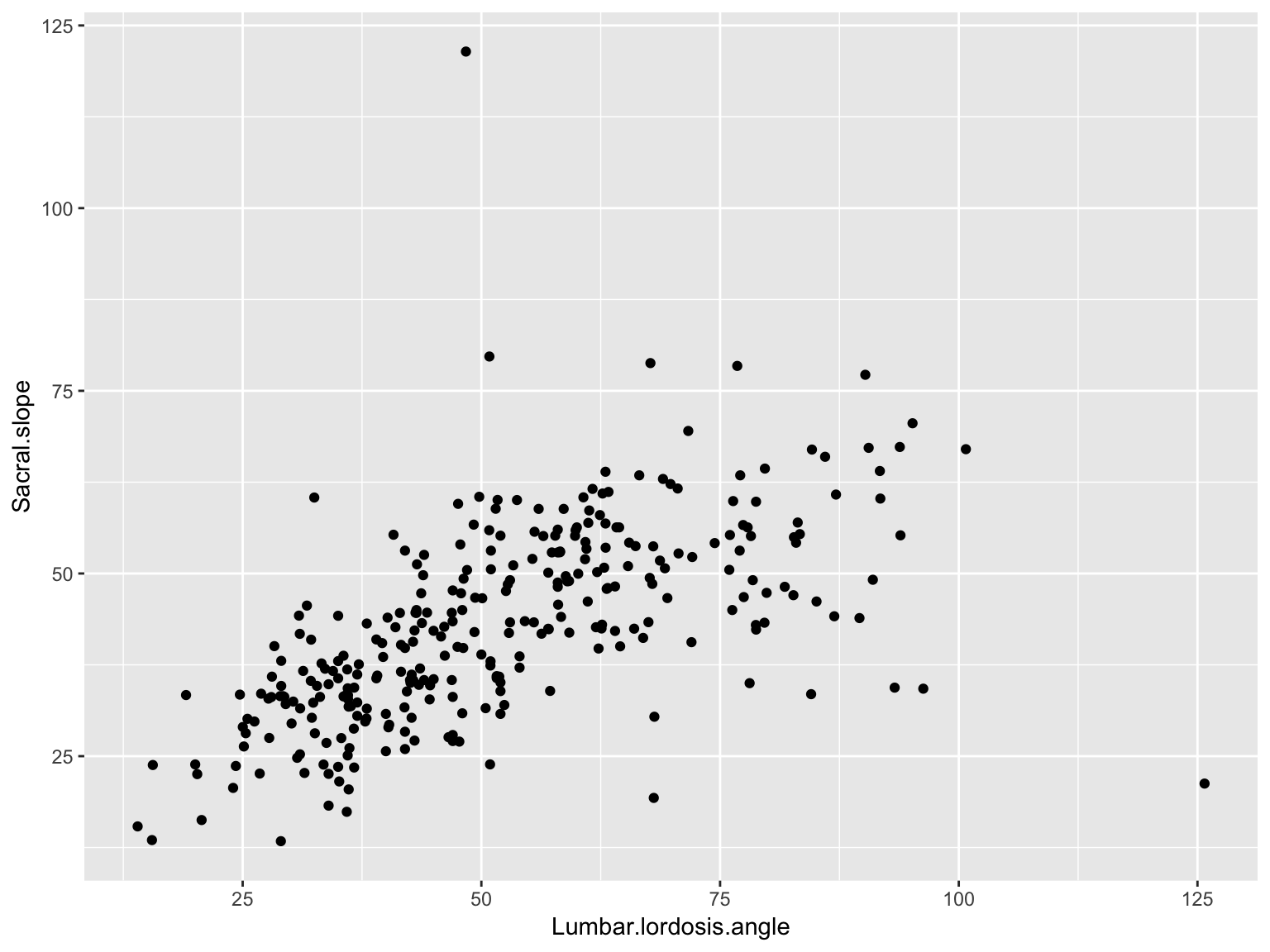
Scatter plot with a smooth line
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope))+
geom_point() +
geom_smooth()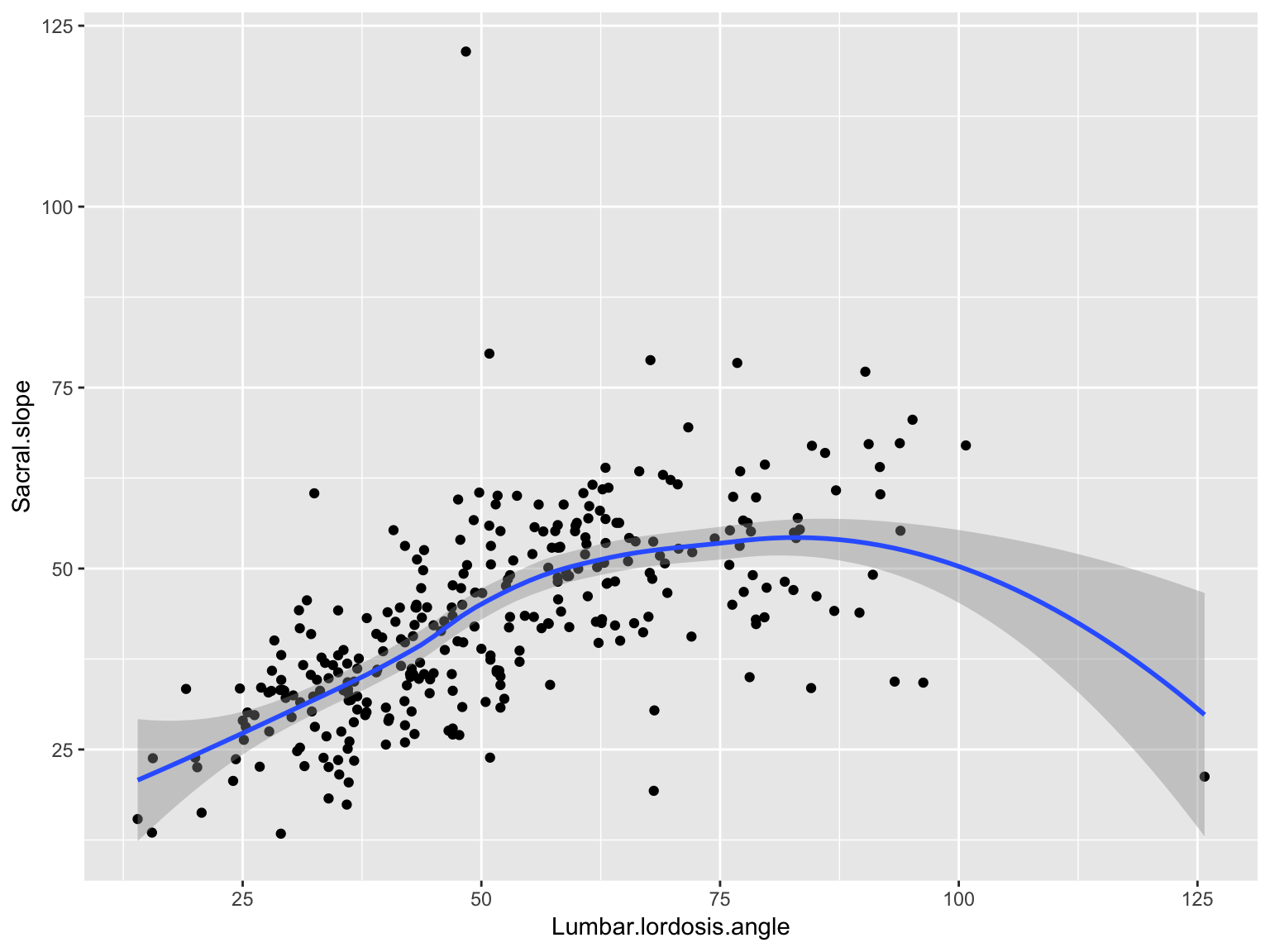
Scatter plot with a smooth straight line
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope)) +
geom_point()+
geom_smooth(method='lm')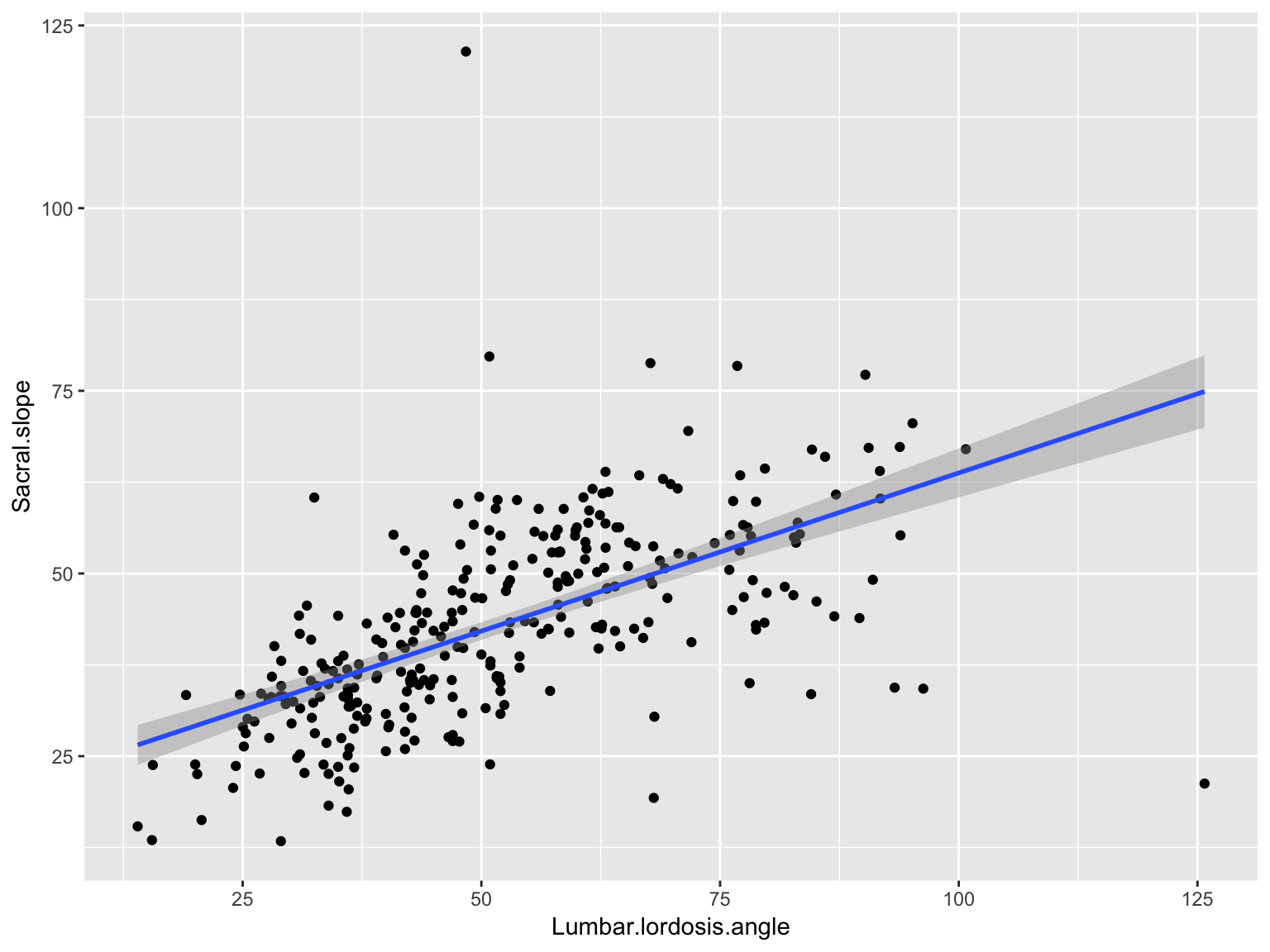
Line plot (for time series)
library(forecast)
d <- data.frame(x = 1:length(gas), y = gas) # Australian monthly gas production
ggplot(d, aes(x, y)) + geom_line()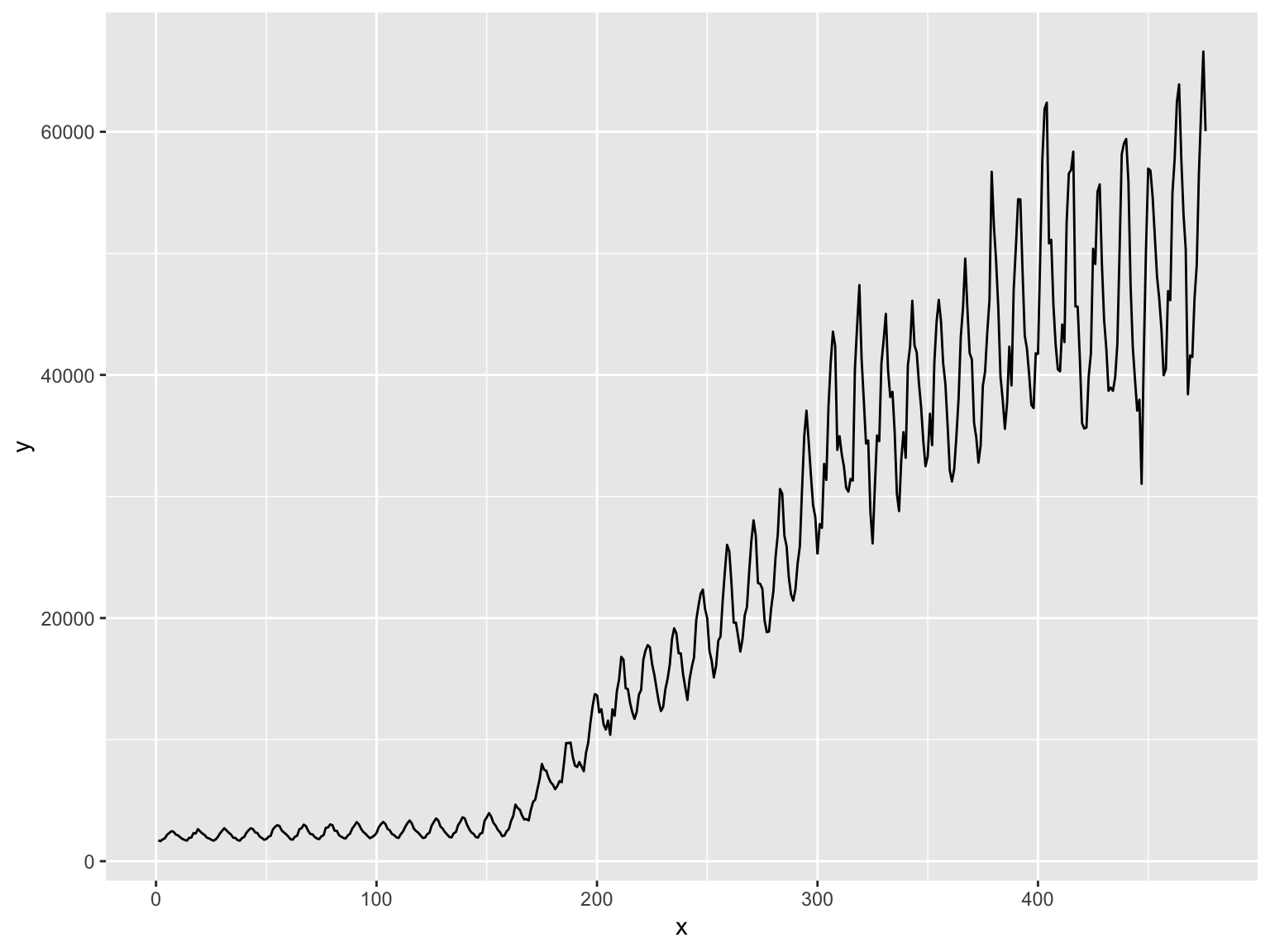
Exercise
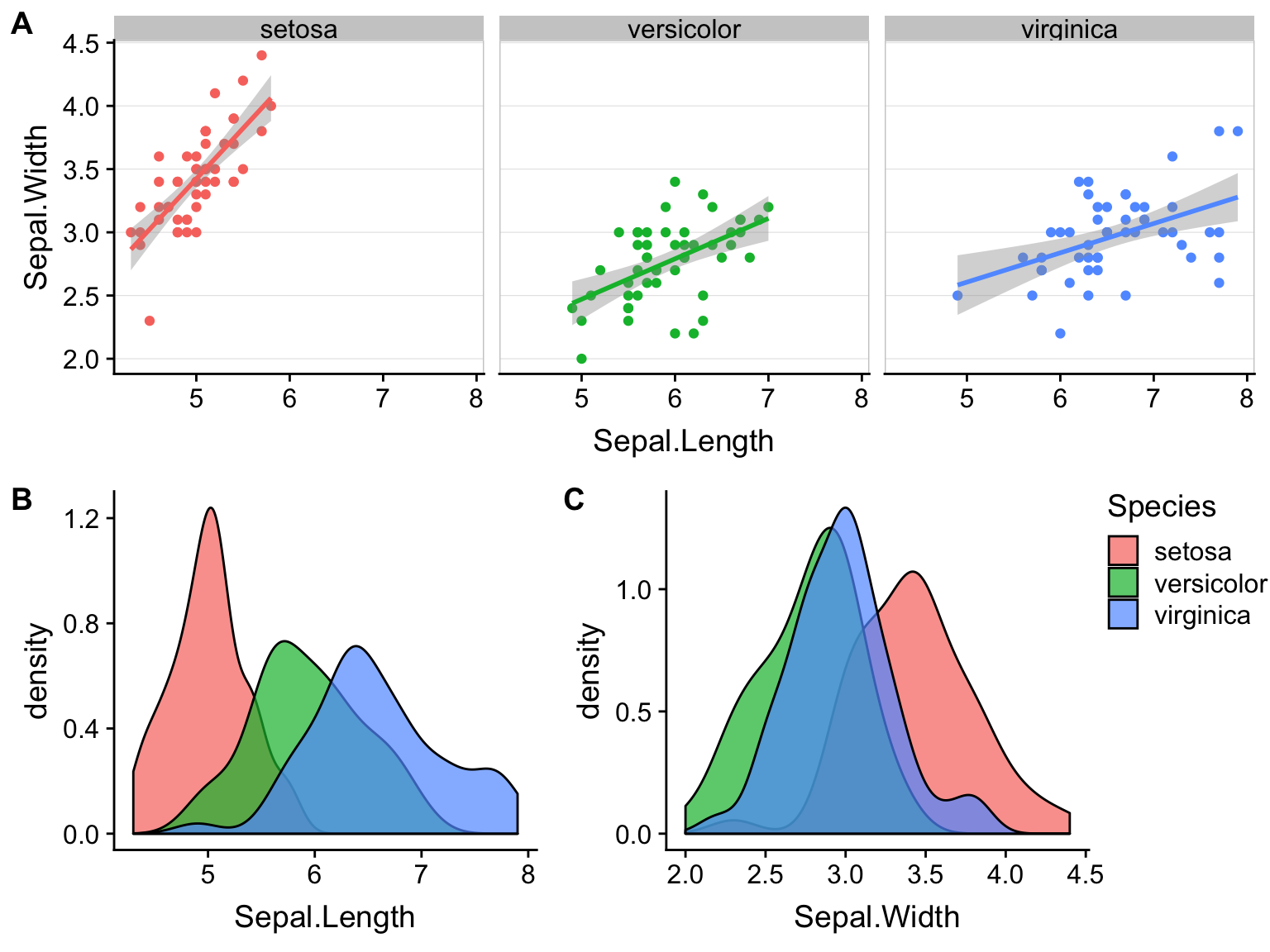
Exercise
- Create a scatter plot of sepal length and sepal width from the
irisdataset, and add a smooth line through it
Solution
ggplot(iris, aes(Sepal.Length, Sepal.Width)) + geom_point() + geom_smooth()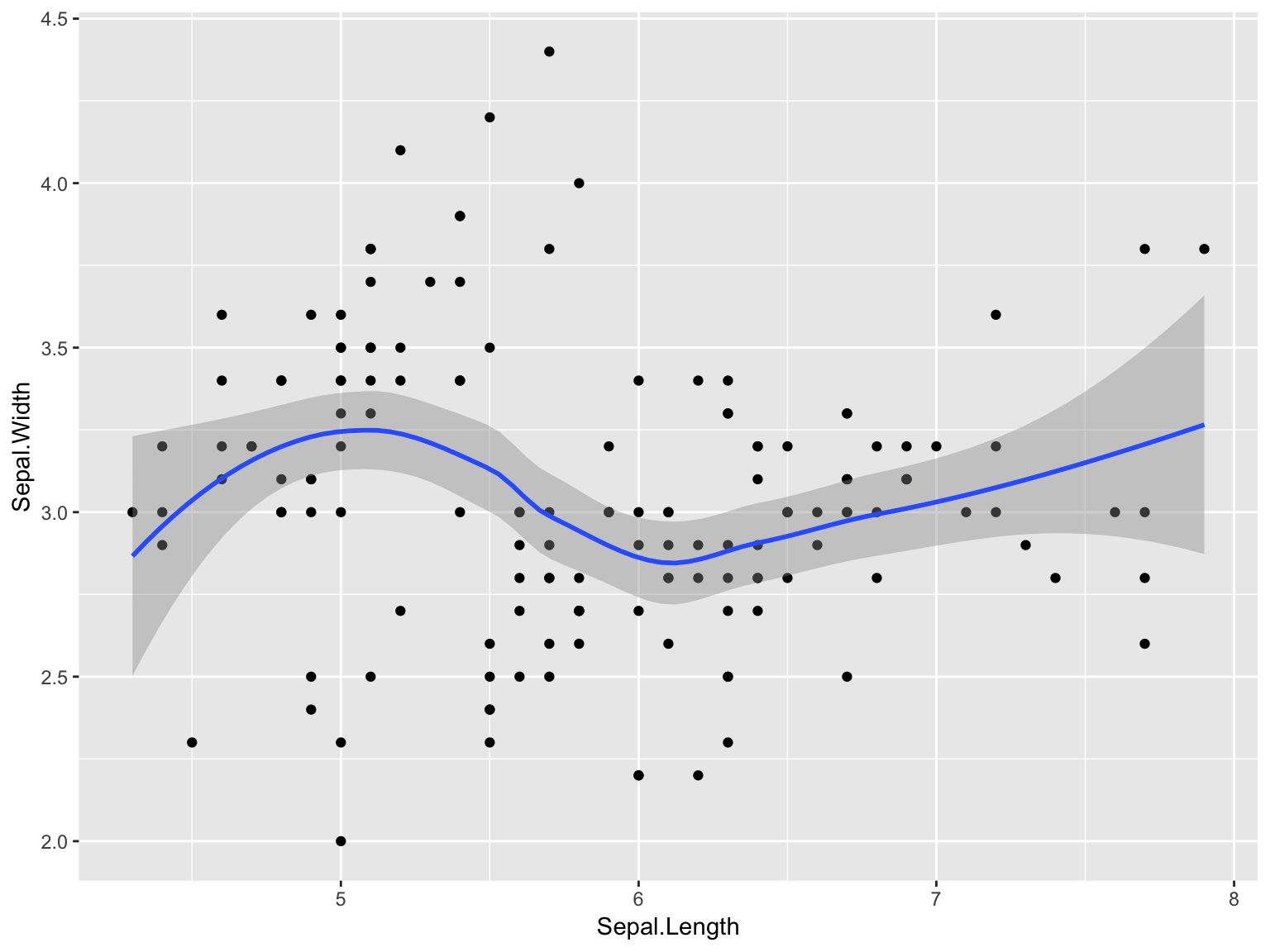
Continuous variable with discrete variable
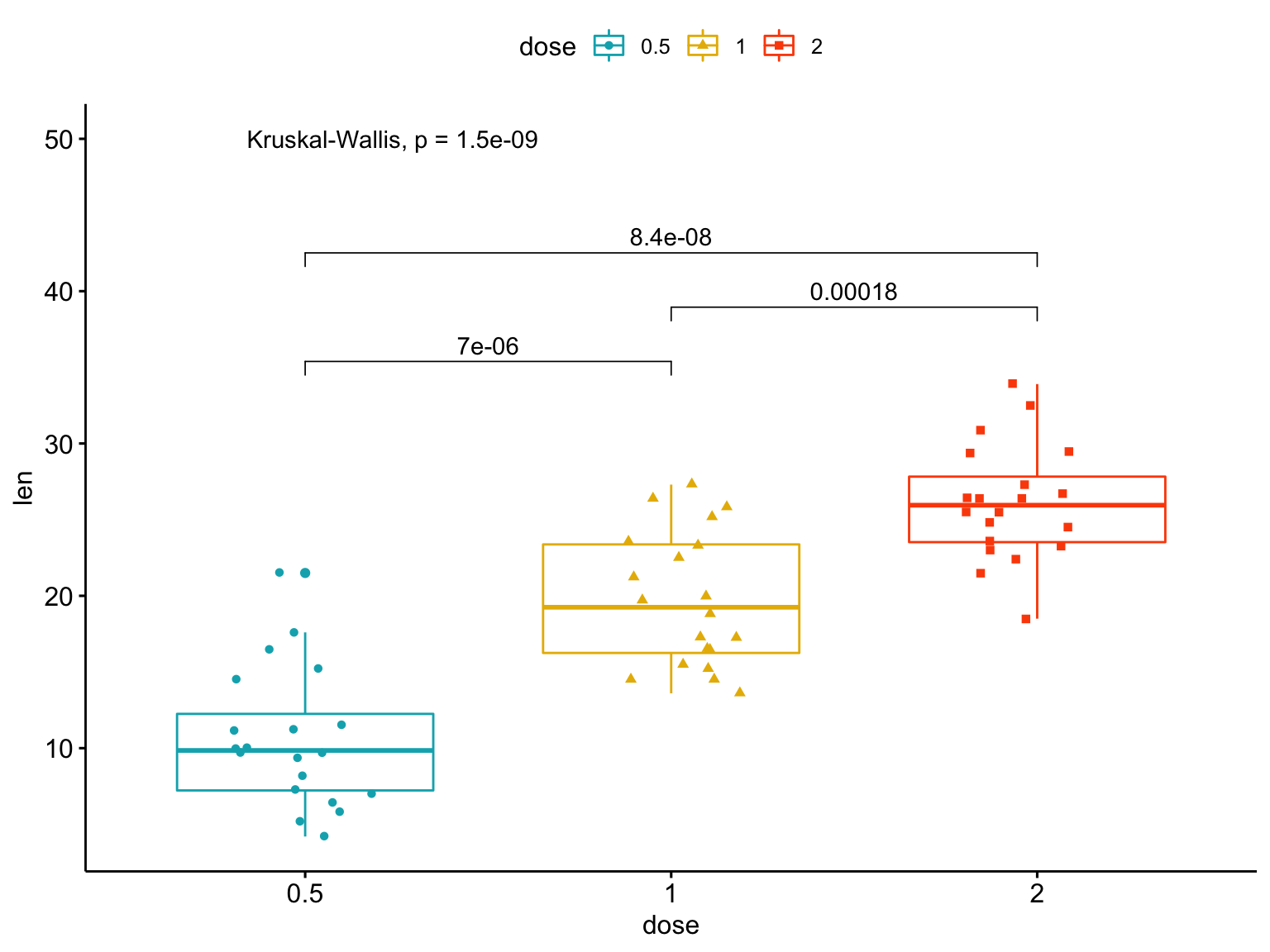
Boxplots
ggplot(data_spine, aes(x = Class.attribute, y = Sacral.slope))+
geom_boxplot()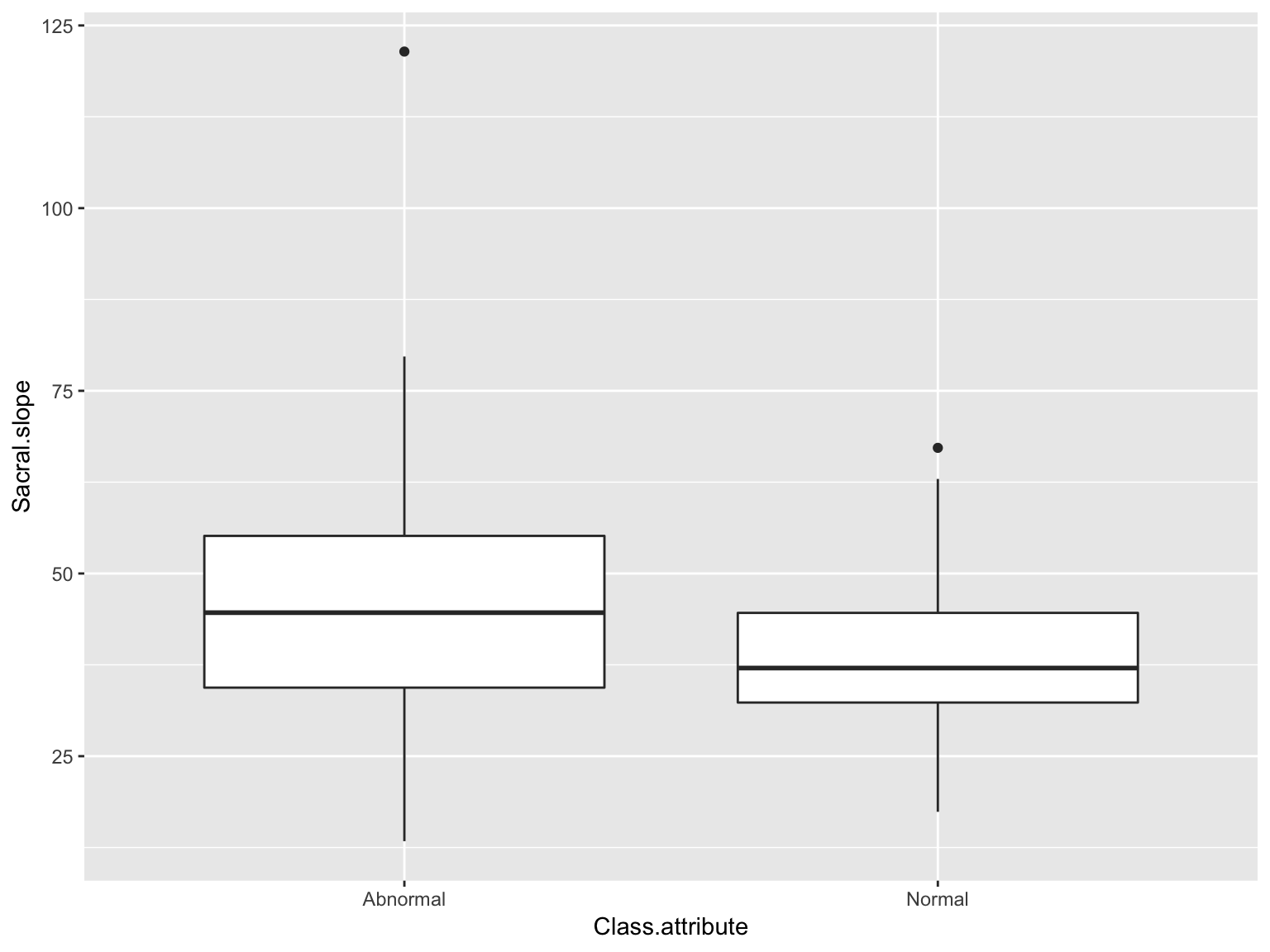
# Factor/discrete variable is always xViolin plots
ggplot(data_spine, aes(x = Class.attribute, y = Sacral.slope)) +
geom_violin()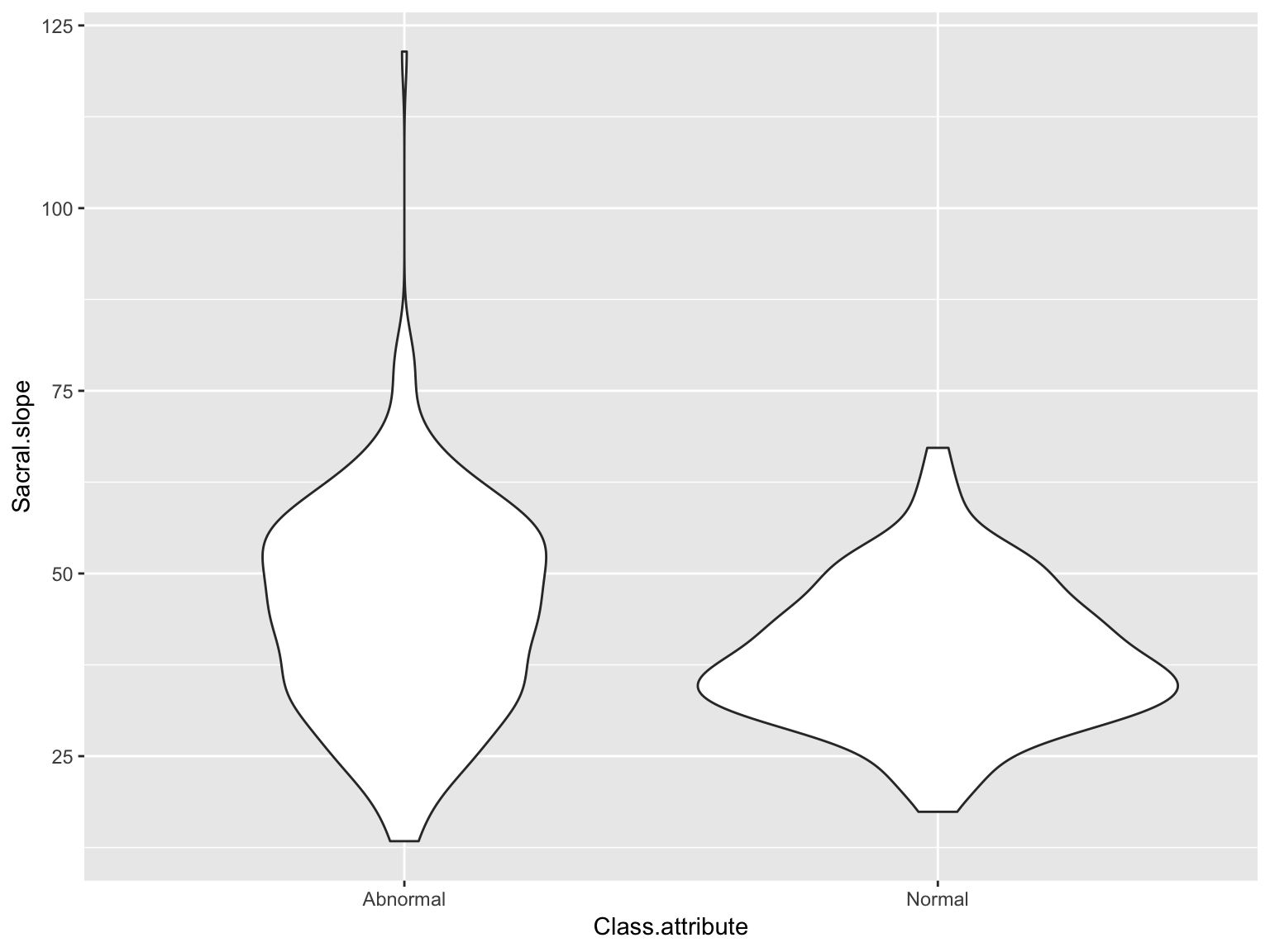
Exercise
Exercise
- Plot a boxplot of petal length by species using the
irisdataset
Solution
ggplot(iris, aes(x = Species, y = Petal.Length))+geom_boxplot()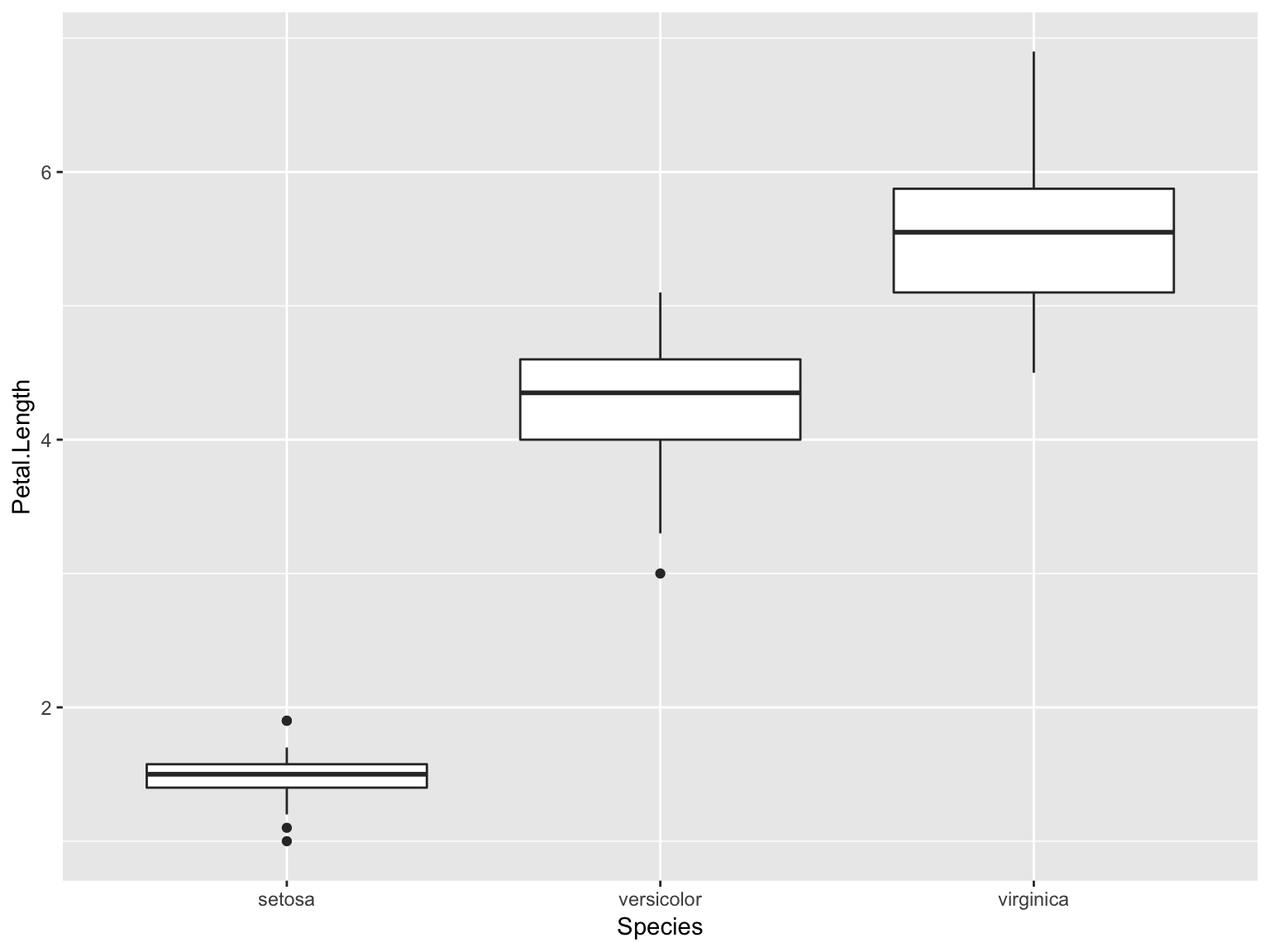
Flipping axes
Vertical bars
ggplot(data_brca, aes(x = Tumor))+geom_bar()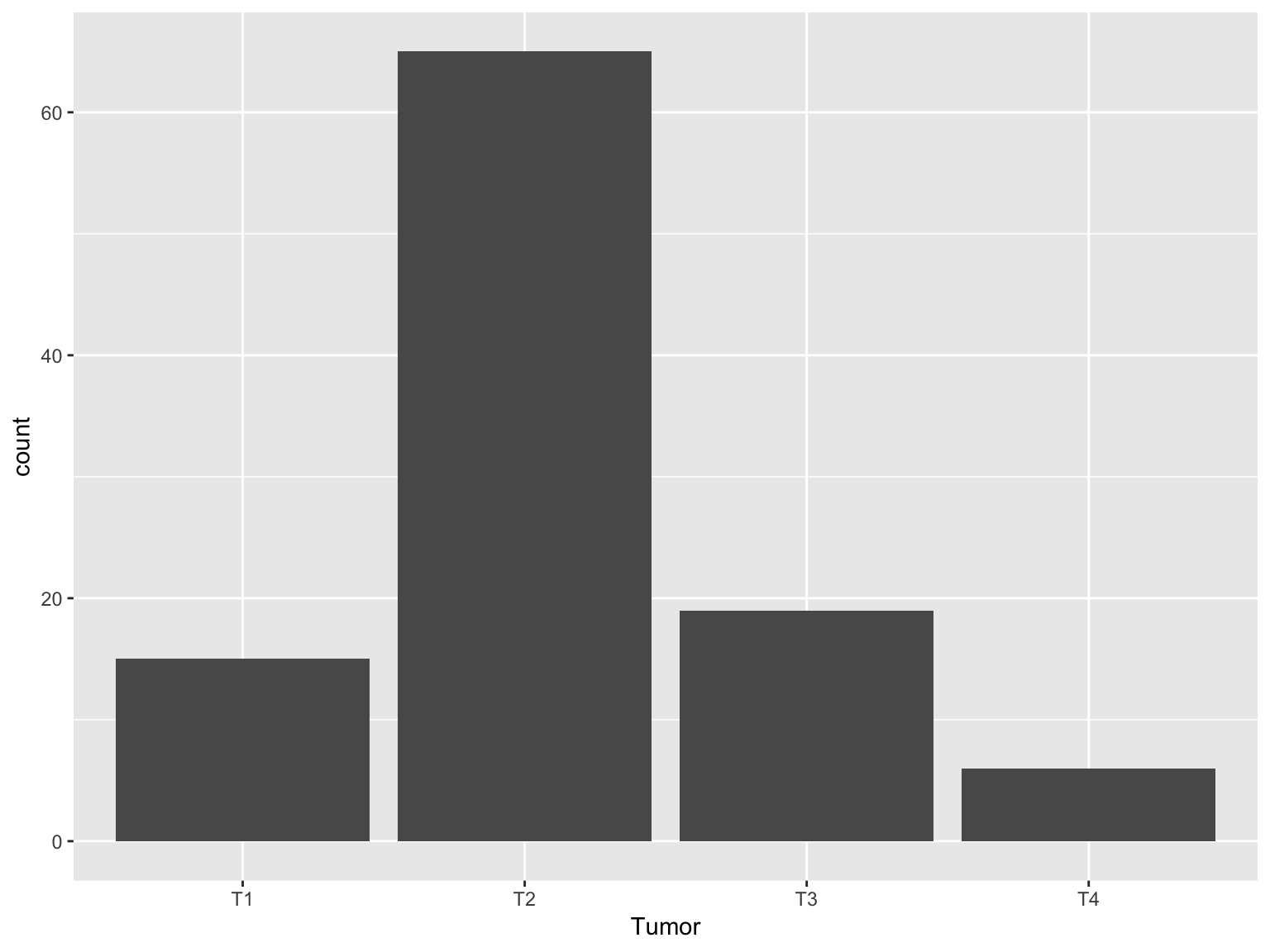
Horizontal bars
ggplot(data_brca, aes(x = Tumor))+geom_bar()+
coord_flip()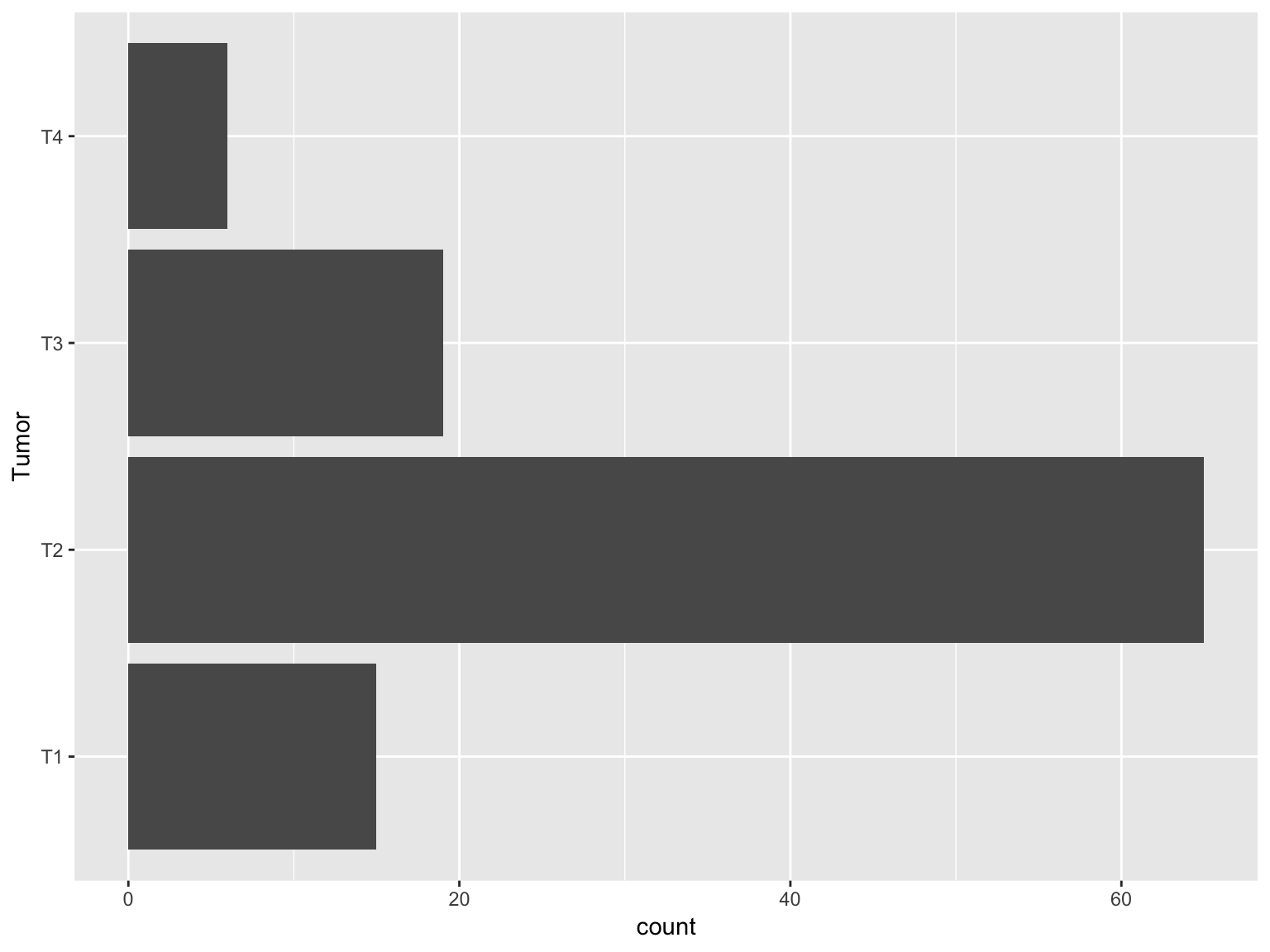
Resources
Online resources
- The ggplot website has many resources to help create visualizations
- The R Graph Gallery
- There are a lot of blogs showing many capabilities of ggplot2
- StackOverflow is the place for Q & A.
Other packages
Group-wise descriptives and visualizations
Grouping
- It is common to look at statistics within subgroups of the data
- The idea is to see if secondary variables affect your primary outcome or relationship
Introducing the dplyr package
dplyr is the most lucid package for manipulating and analyzing data organized in a data frame.
- It has a
group_byfunction which creates a grouped data frame
library(dplyr)
grouped_data_spine = data_spine %>% group_by(Class.attribute)Note that you have to group using a discrete valued variable (factor, character, integer)
Grouped summaries
grouped_data_spine %>%
summarize(mean(Pelvic.incidence),
sd(Pelvic.incidence),
min(Pelvic.incidence),
max(Pelvic.incidence))| Class.attribute | mean(Pelvic.incidence) | sd(Pelvic.incidence) | min(Pelvic.incidence) | max(Pelvic.incidence) |
|---|---|---|---|---|
| Abnormal | 64.69 | 17.66 | 26.15 | 129.83 |
| Normal | 51.69 | 12.37 | 30.74 | 89.83 |
Grouped summaries
grouped_data_spine %>% summarize(Mean = mean(Pelvic.incidence),
SD = sd(Pelvic.incidence),
Min = min(Pelvic.incidence),
Max = max(Pelvic.incidence))Grouped summaries
grouped_data_spine %>% summarize_all(mean)# # A tibble: 2 x 13
# Class.attribute Pelvic.incidence Pelvic.tilt Lumbar.lordosis…
# <fct> <dbl> <dbl> <dbl>
# 1 Abnormal 64.7 19.8 55.9
# 2 Normal 51.7 12.8 43.5
# # ... with 9 more variables: Sacral.slope <dbl>, Pelvic.radius <dbl>,
# # Degree.spondylolisthesis <dbl>, Pelvic.slope <dbl>, Direct.tilt <dbl>,
# # Thoracic.slope <dbl>, Cervical.tilt <dbl>, Sacrum.angle <dbl>,
# # Scoliosis.slope <dbl>A note on tibbles
- Tibbles are a new-generation object meant to enhance the
data.frame.- Central to thetidyverse packages
- If you want to just get back to a more familiar
data.frameobject, useas.data.frame - A tibble is built on a
data.frame, so all operations ondata.frame’s will work. - To see all columns, set
options(dplyr.width=Inf).
A note on tibbles
Differences between a tibble and a data.frame:
- Printing a tibble is restricted to the first 10 lines, and includes column types
- Stricter subsetting rules that make the types of objects created consistent
Using ggplot in a pipeline
data_spine %>%
group_by(Class.attribute) %>%
summarize_all(funs(Mean = mean(., na.rm=T),
SEM = sd(., na.rm=T)/sqrt(n()))) %>%
gather(variable, value, -Class.attribute) %>%
separate(variable, c('Variable','stat'), sep = '_') %>%
spread(stat, value) %>%
mutate(lcb = Mean - 2 * SEM, ucb = Mean + 2 * SEM) %>%
ggplot(aes(x = Class.attribute, y = Mean, ymin = lcb, ymax = ucb)) +
geom_pointrange() +
facet_wrap(~Variable, scales = 'free_y') +
labs( x = 'Class', y = '') +
ggtitle('Confidence intervals of the mean')Work through the pipeline yourself to understand what each step does, just like last week
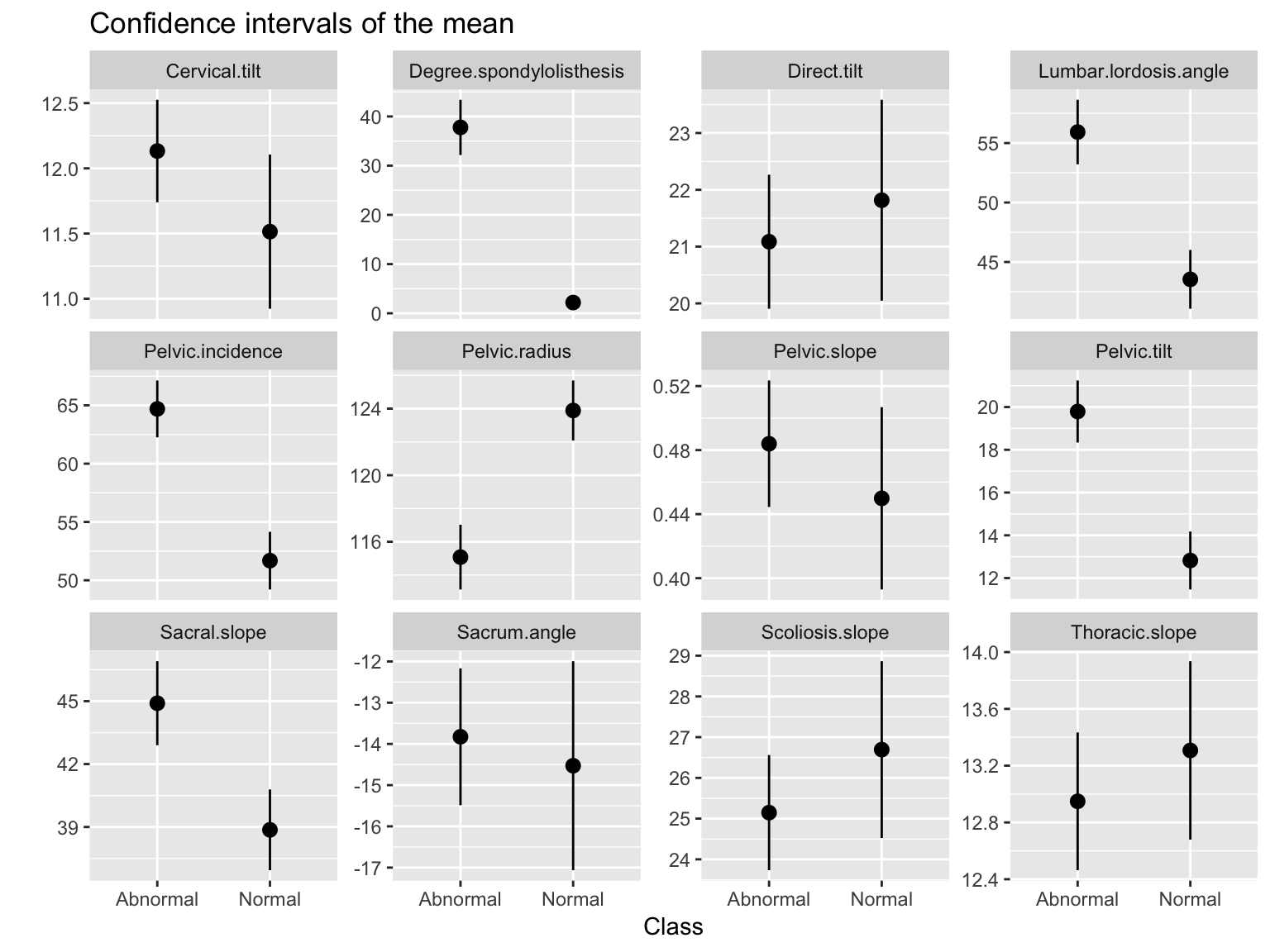
Grouped visualization
Density plot
ggplot(data_spine, aes(x = Sacral.slope, group = Class.attribute,
color=Class.attribute))+
geom_density()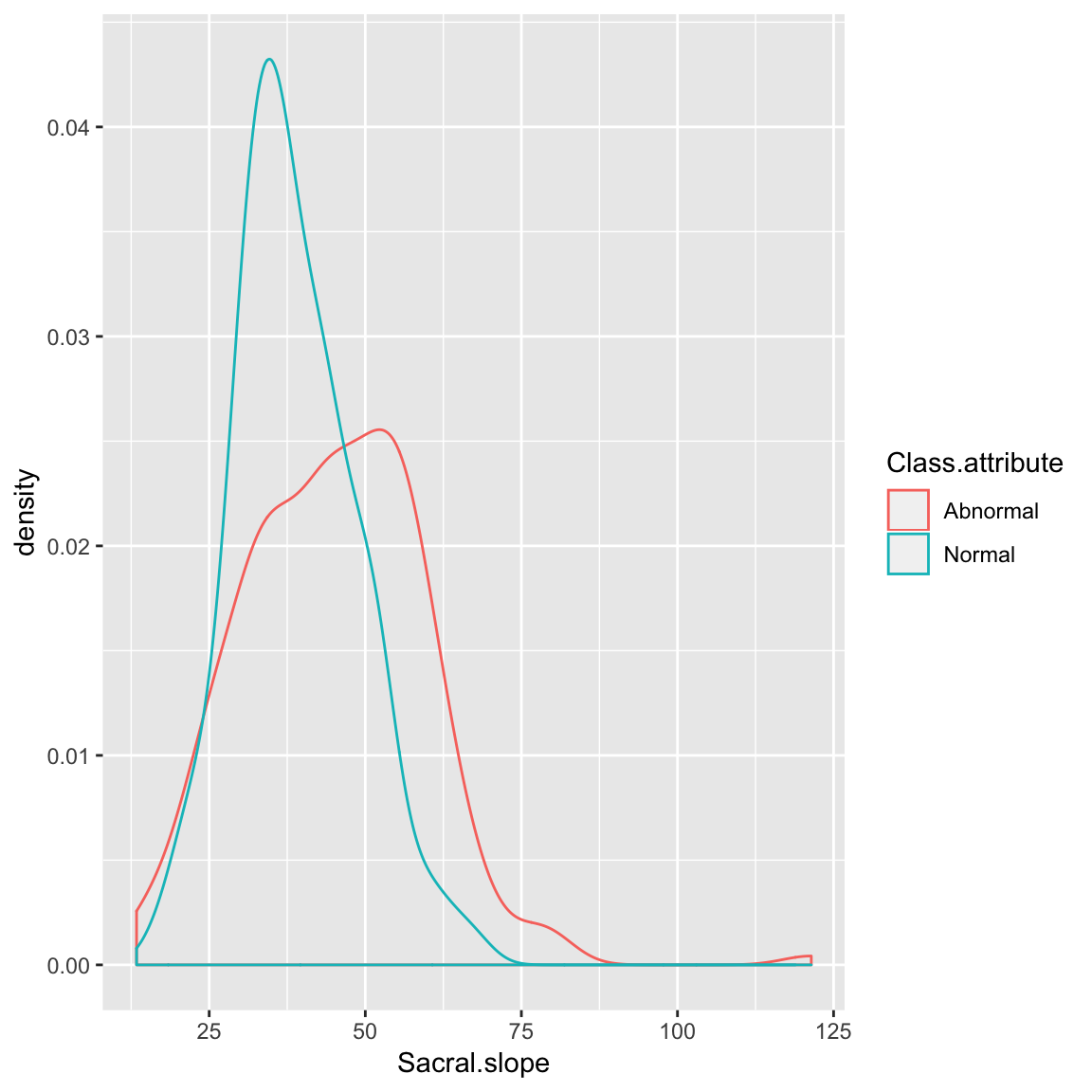
Scatter plot
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope,
group = Class.attribute, color = Class.attribute))+
geom_point()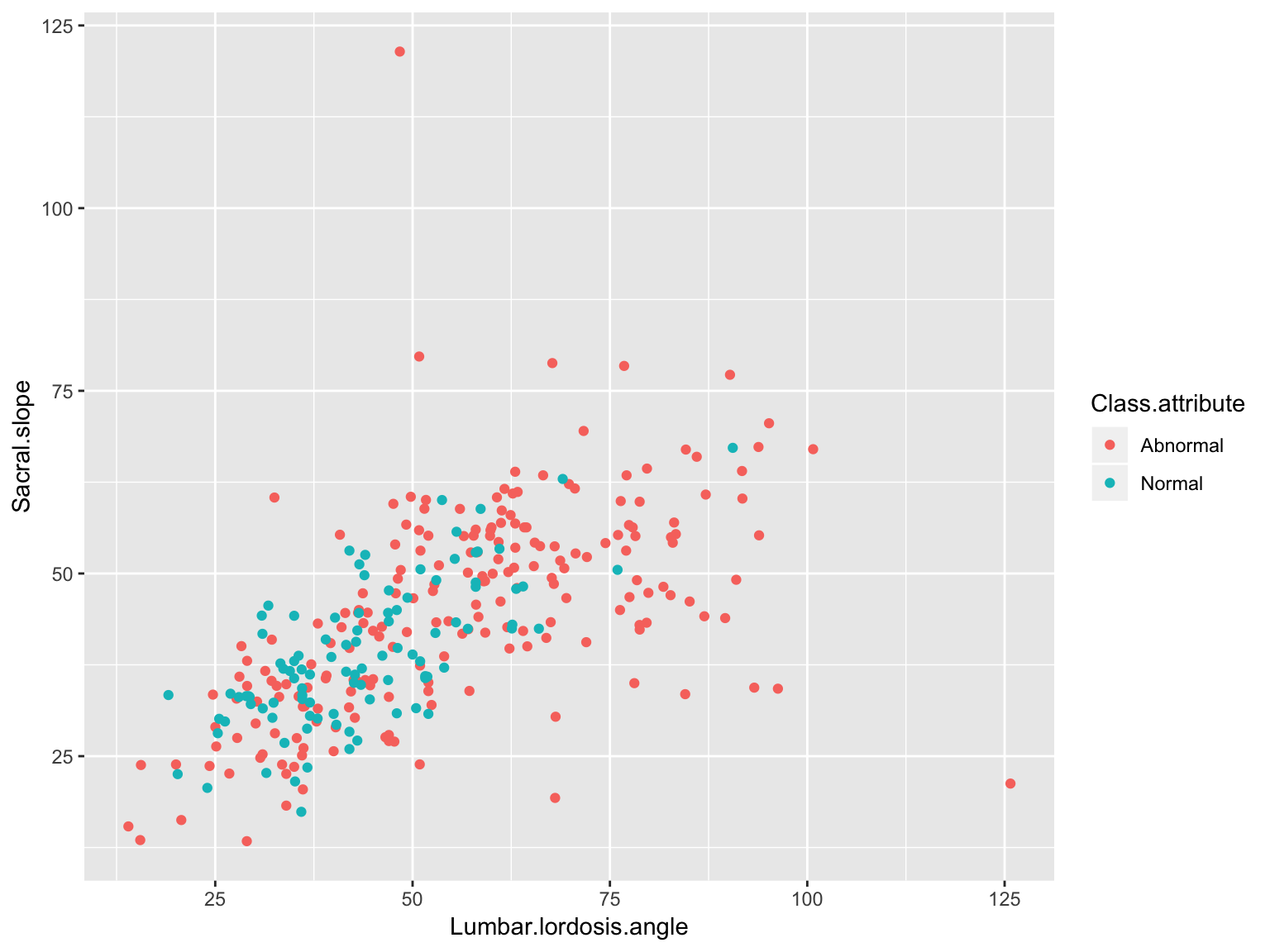
Scatter plot (Black and White)
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope,
group = Class.attribute, shape = Class.attribute))+
geom_point()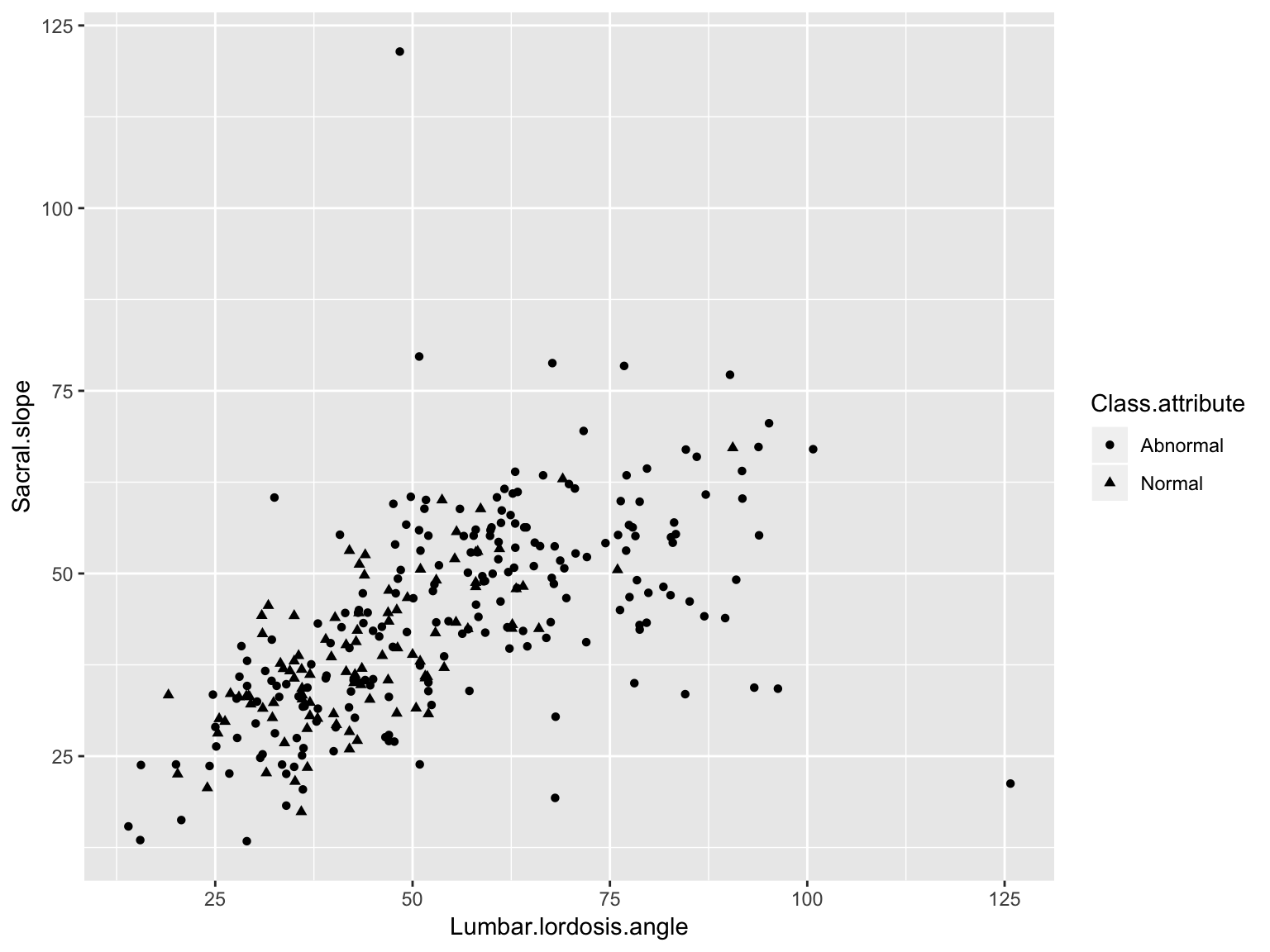
Scatter plot with size representing third variable
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope))+
geom_point(aes(size = Pelvic.slope))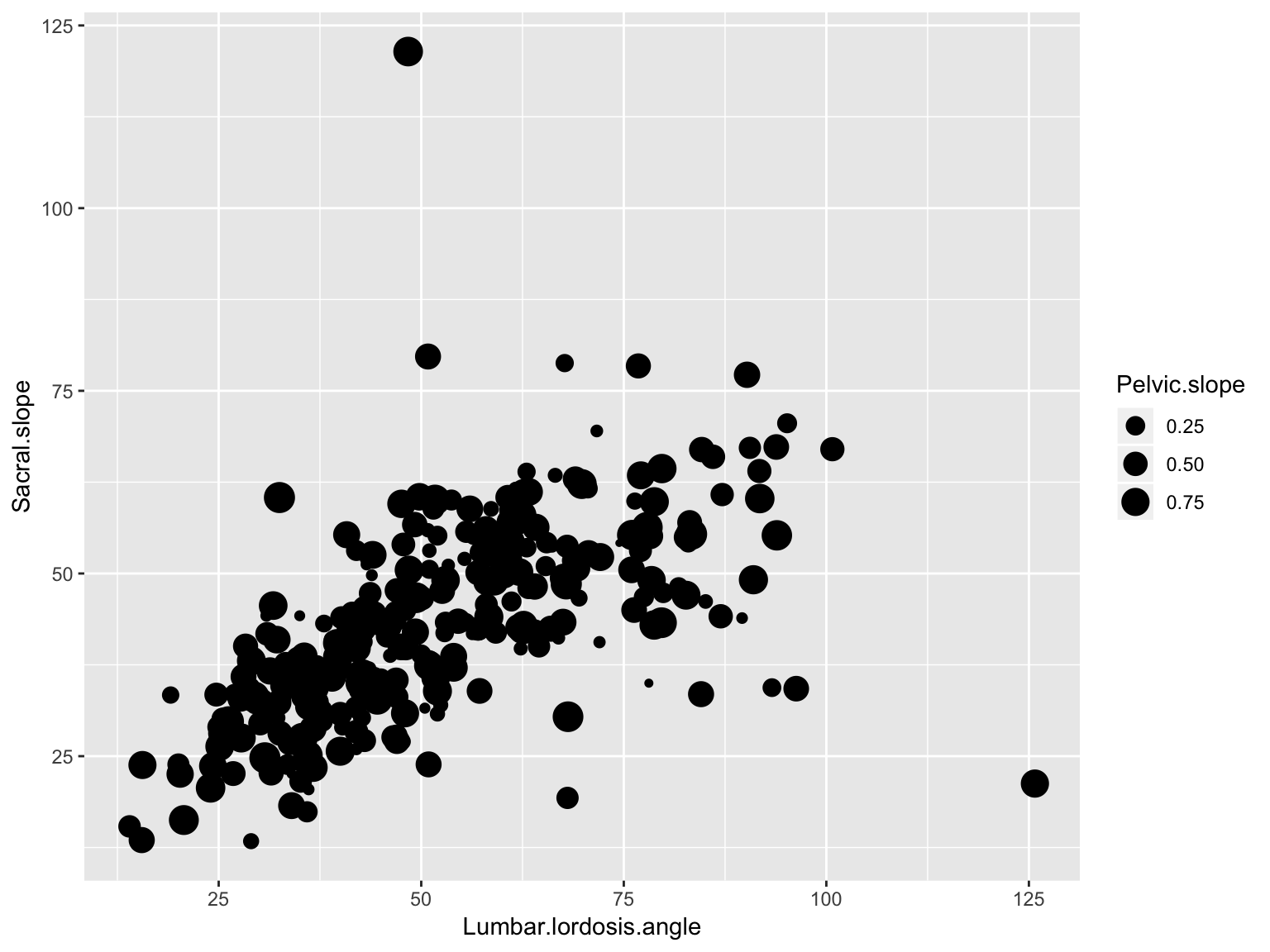
Scatter plot combinations
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope,
group = Class.attribute, color = Class.attribute))+
geom_point(aes(size = Pelvic.slope))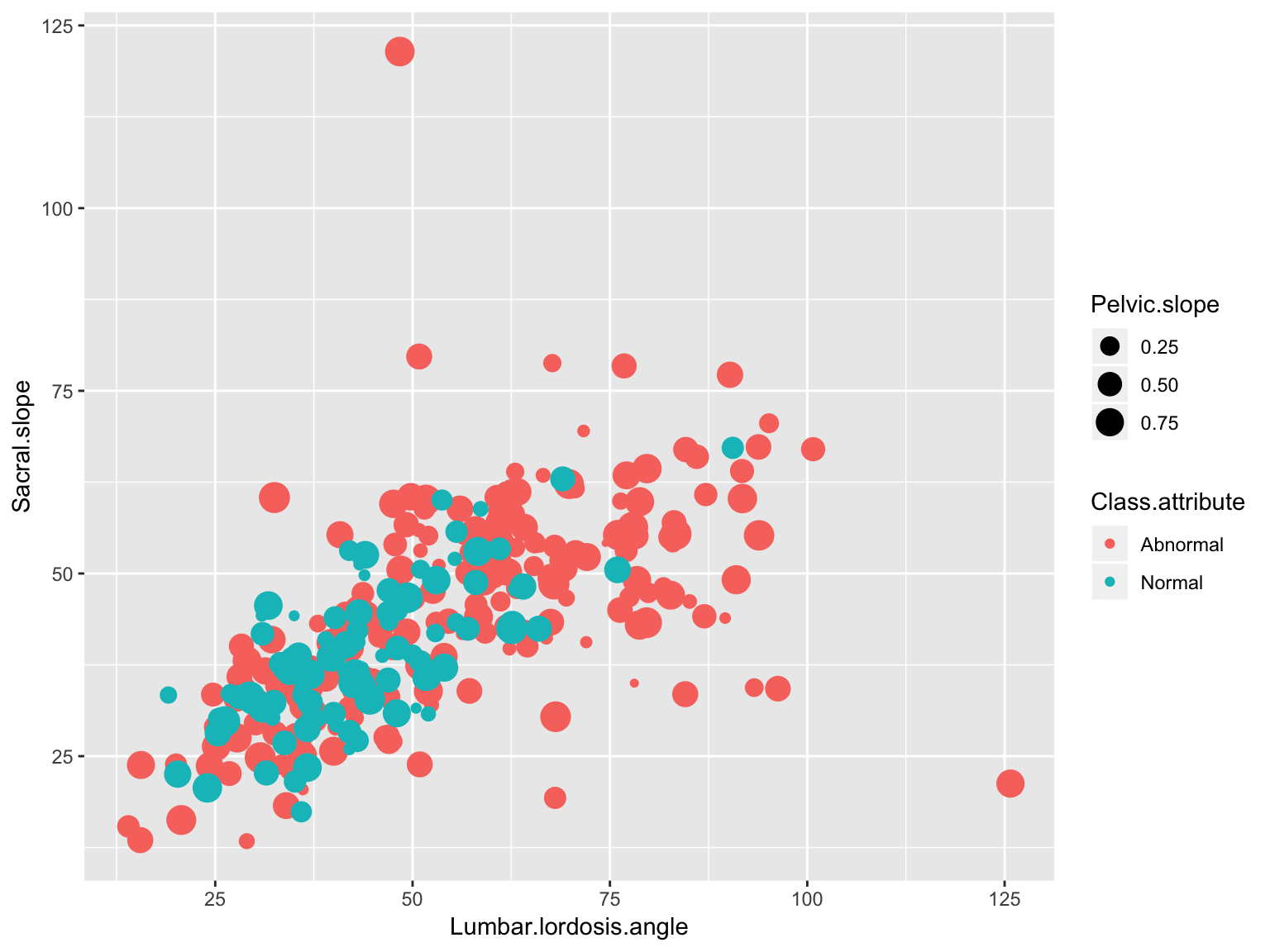
Scatter plot with lines
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope,
group = Class.attribute, color=Class.attribute))+
geom_point()+
geom_smooth(method='lm')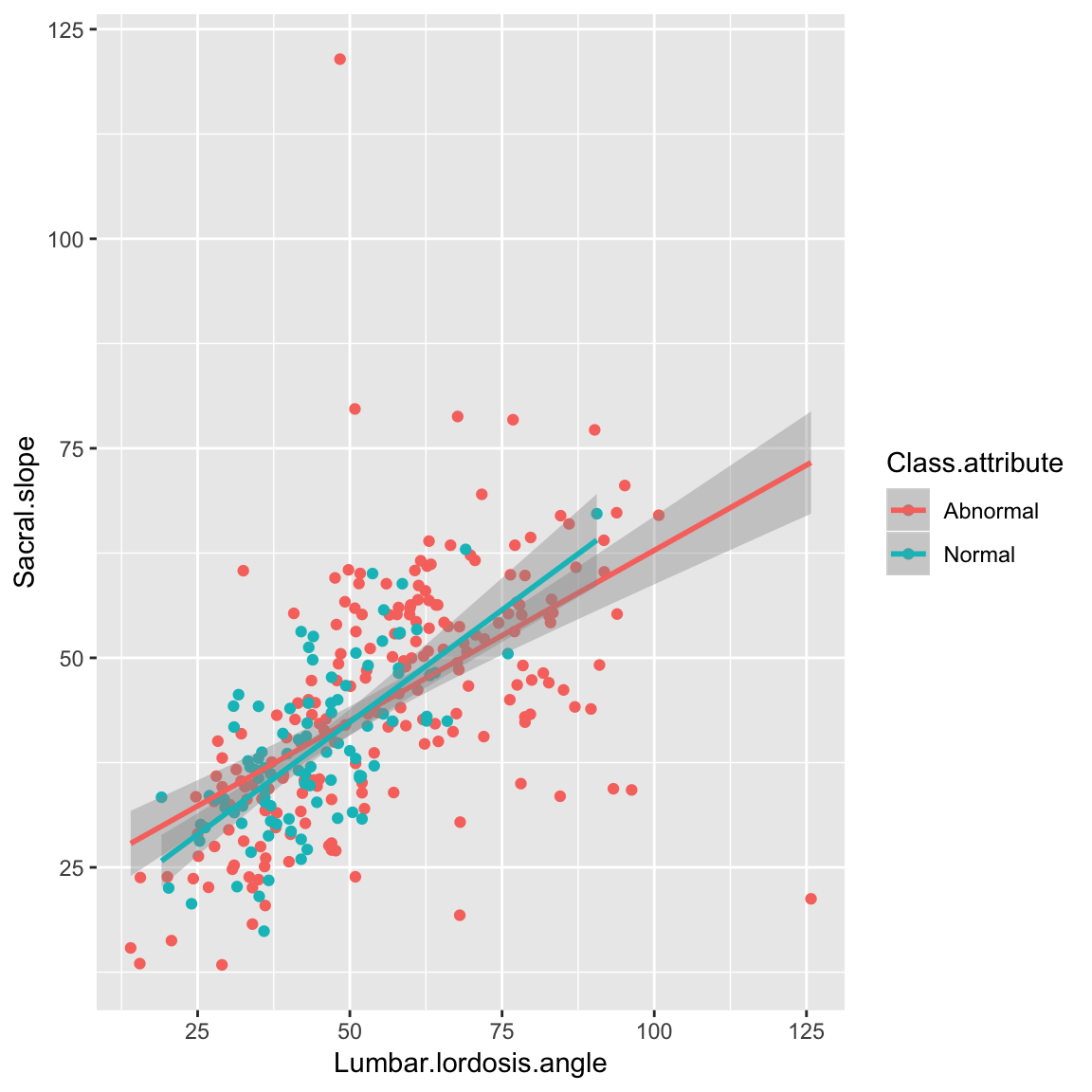
Scatter plot with lines
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope))+
geom_point()+
geom_smooth(aes(color = Class.attribute), method='lm')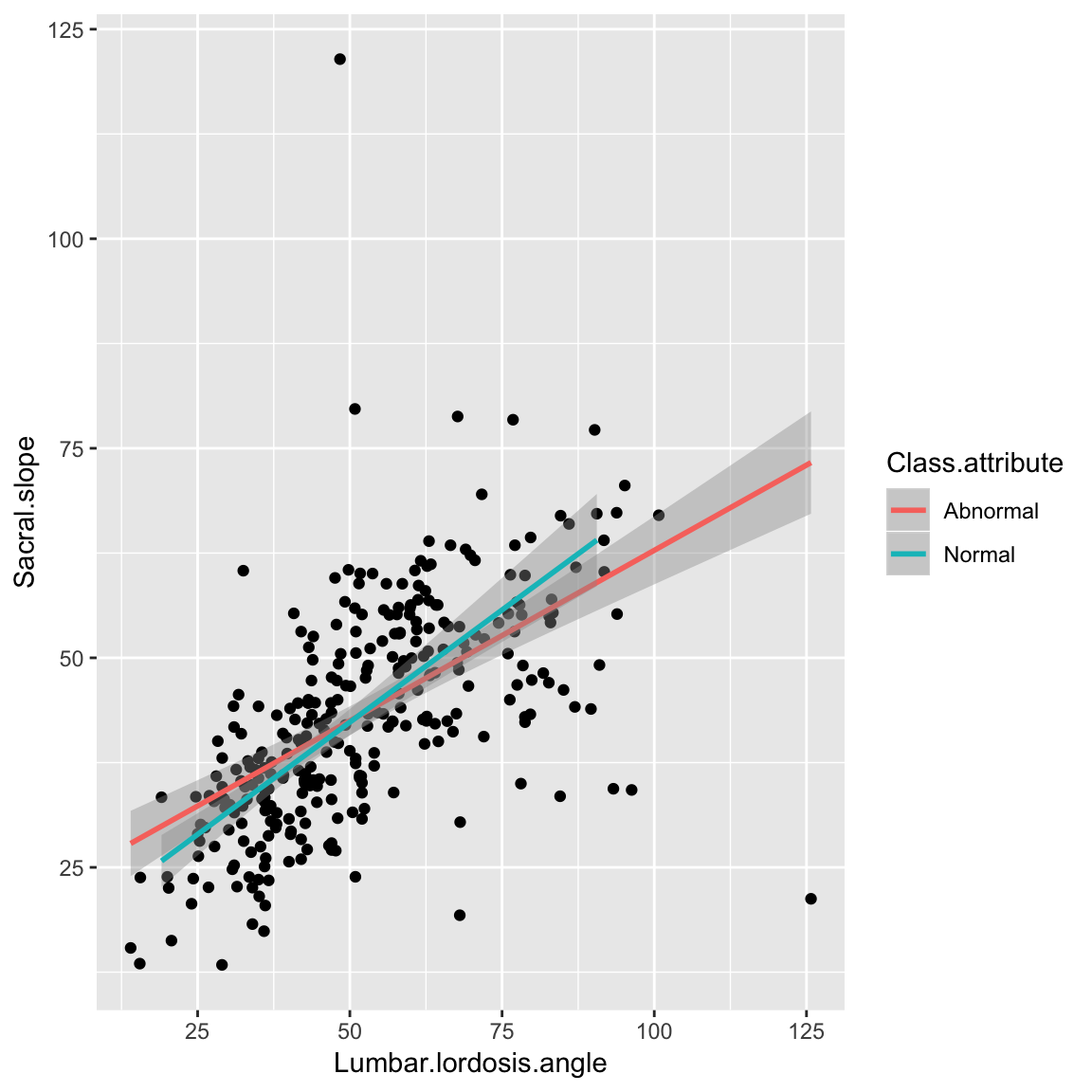
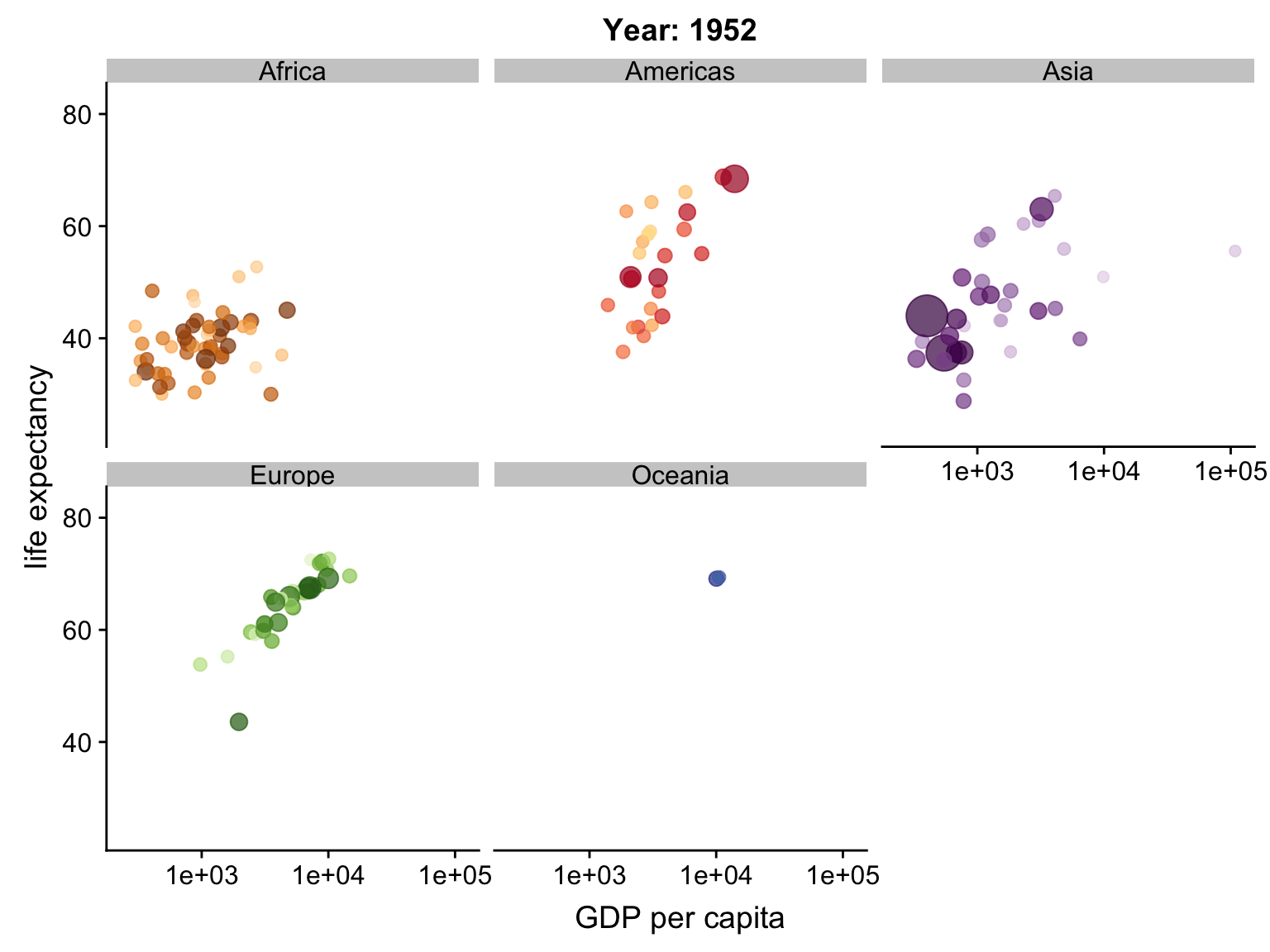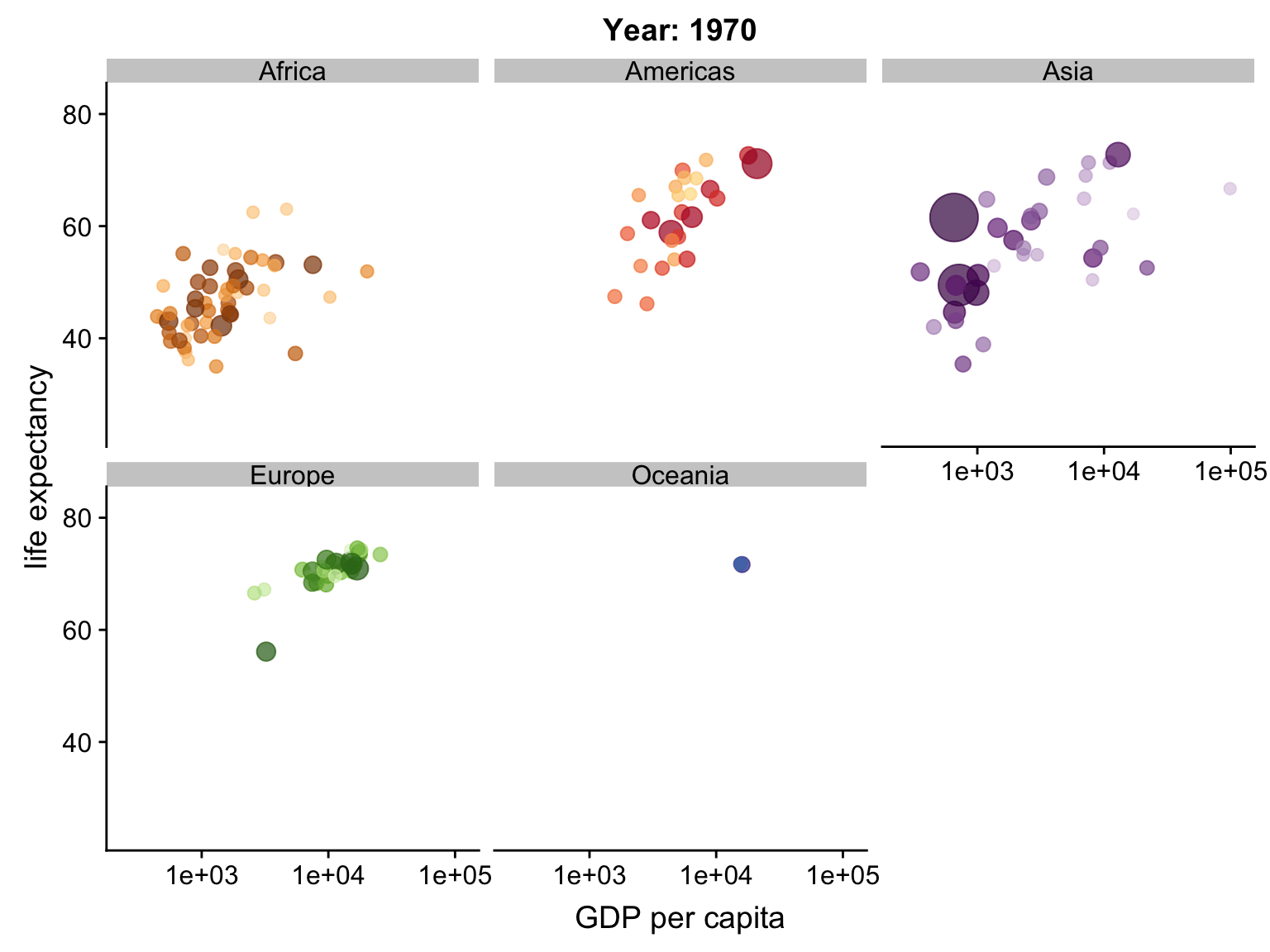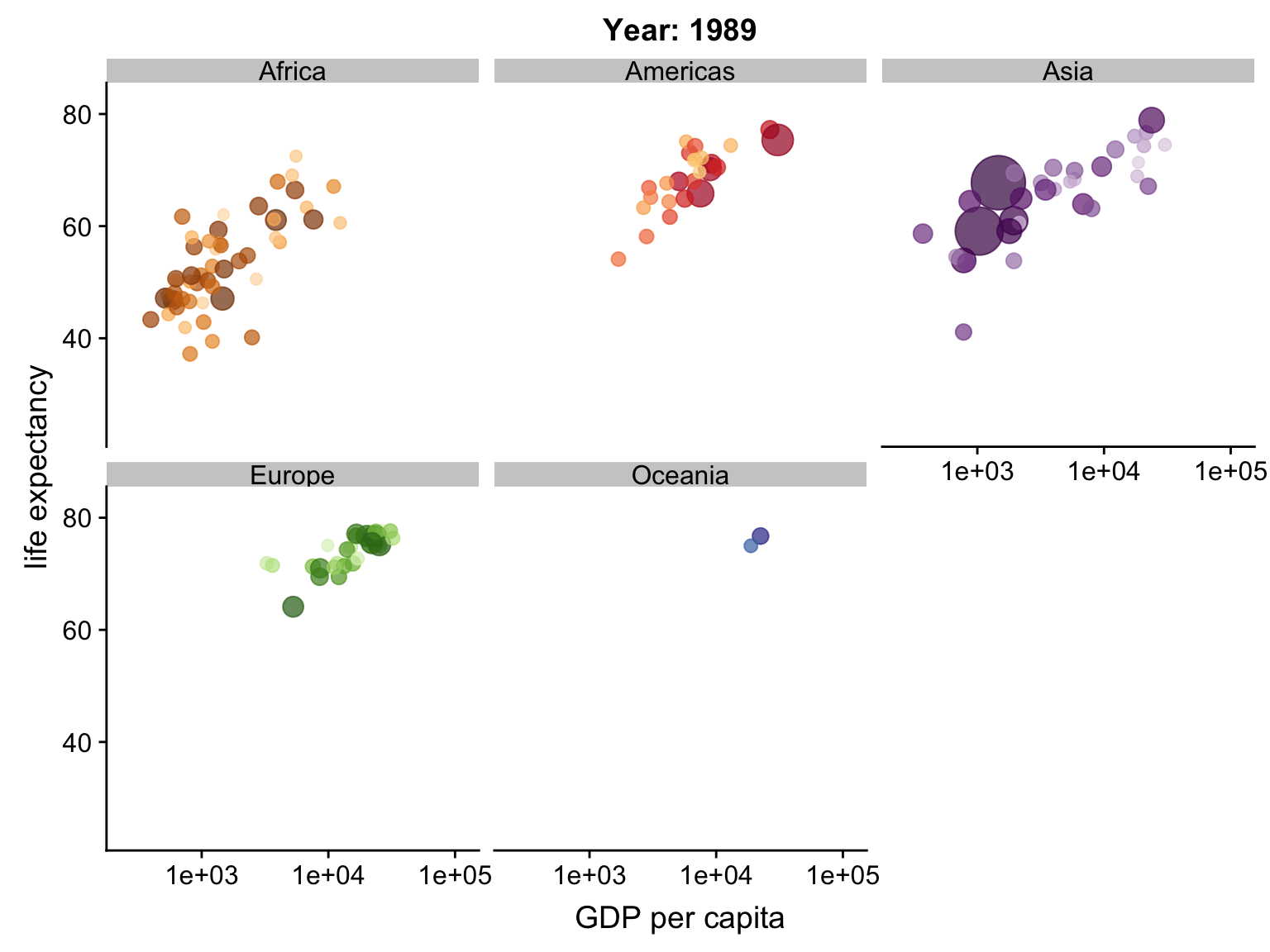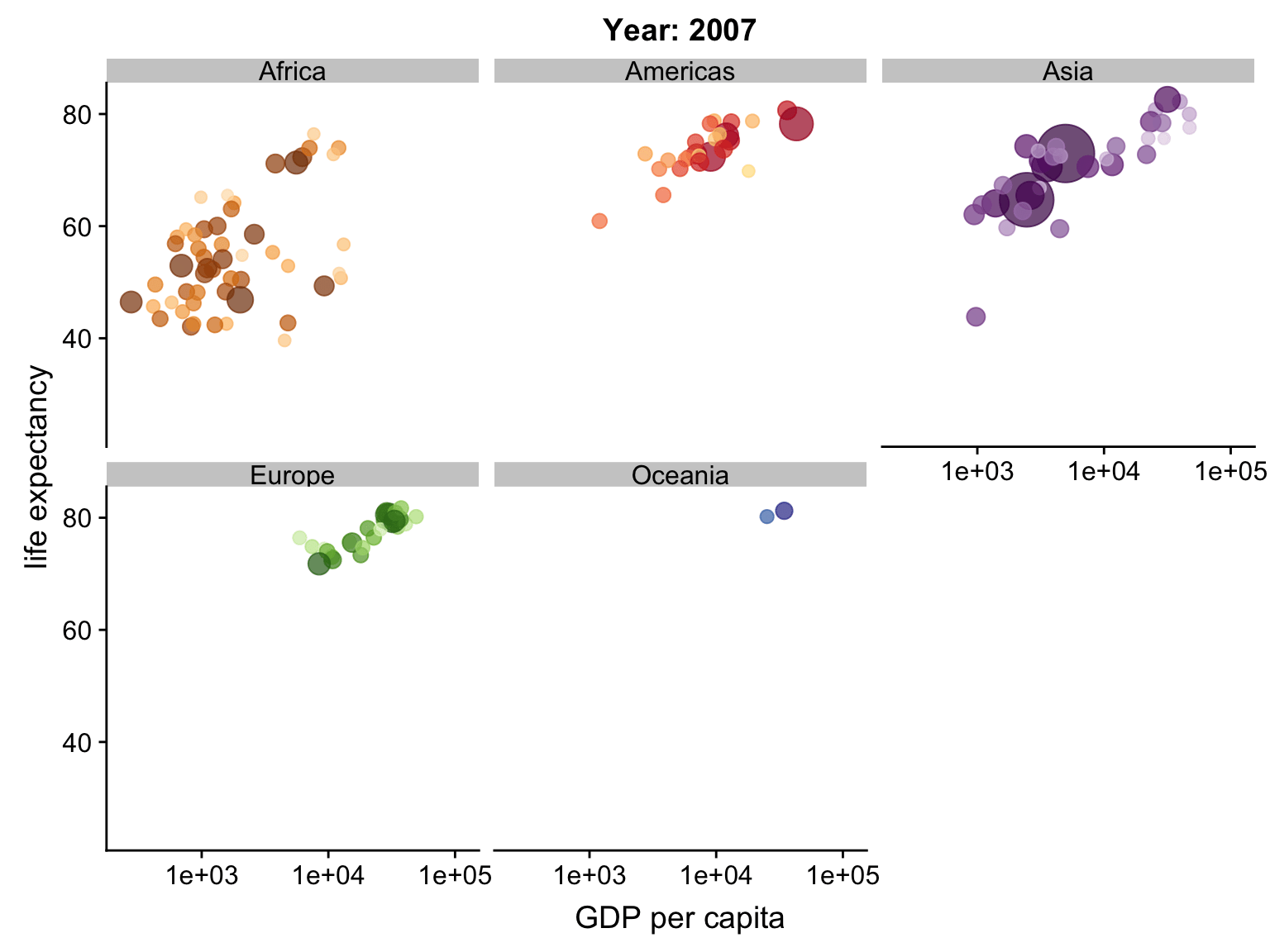
Facetting
Facetting
Facetted graphs are a panel of graphs, each of which corresponds to a particular subgroup of the data.
Facetted scatter plot
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope))+
geom_point()+
facet_wrap( ~ Class.attribute, nrow=1)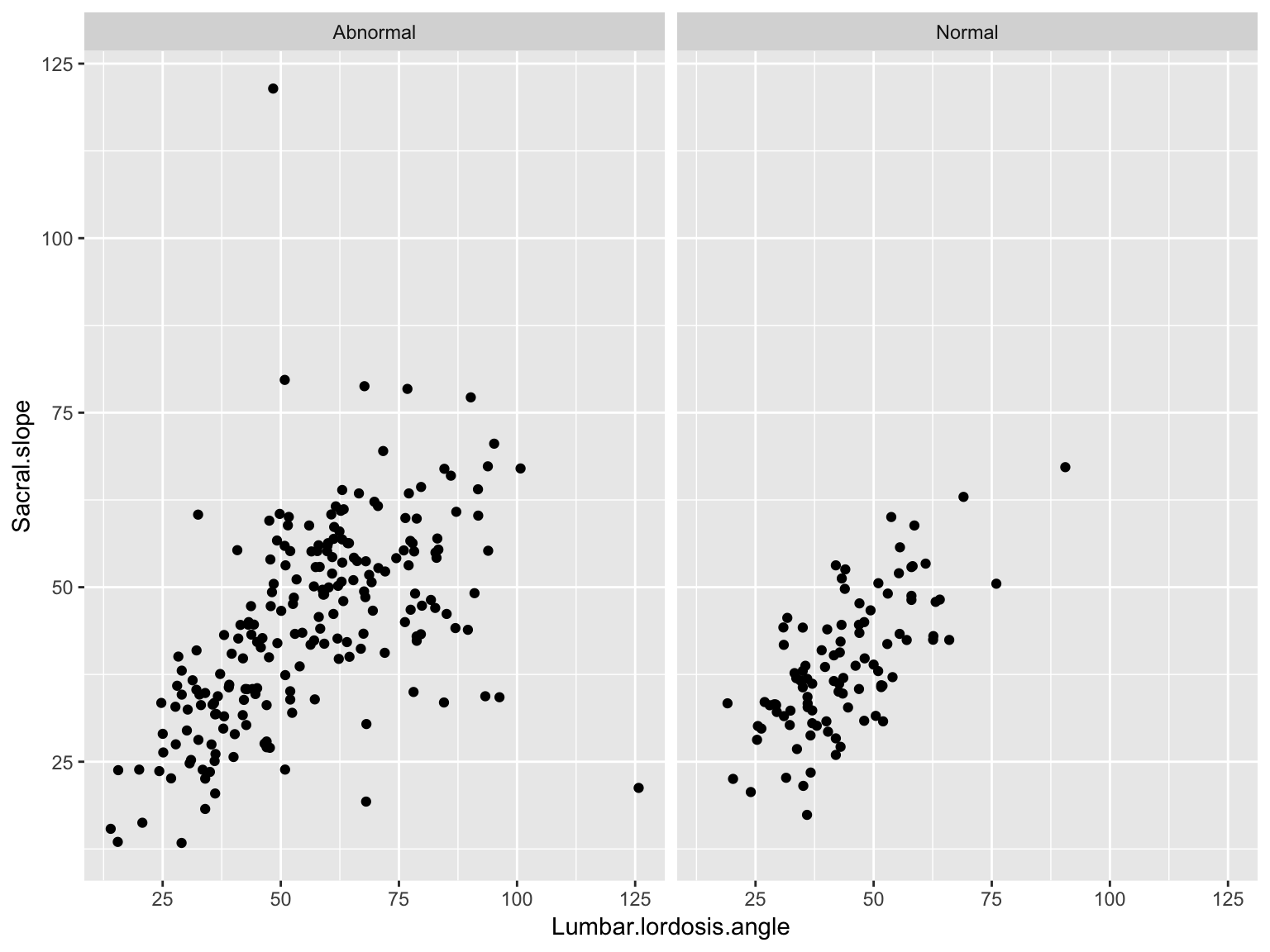
Facetted scatter plot with lines
ggplot(data_spine, aes(x = Lumbar.lordosis.angle, y = Sacral.slope))+
geom_point()+ geom_smooth(method='lm')+
facet_wrap( ~ Class.attribute, nrow=1)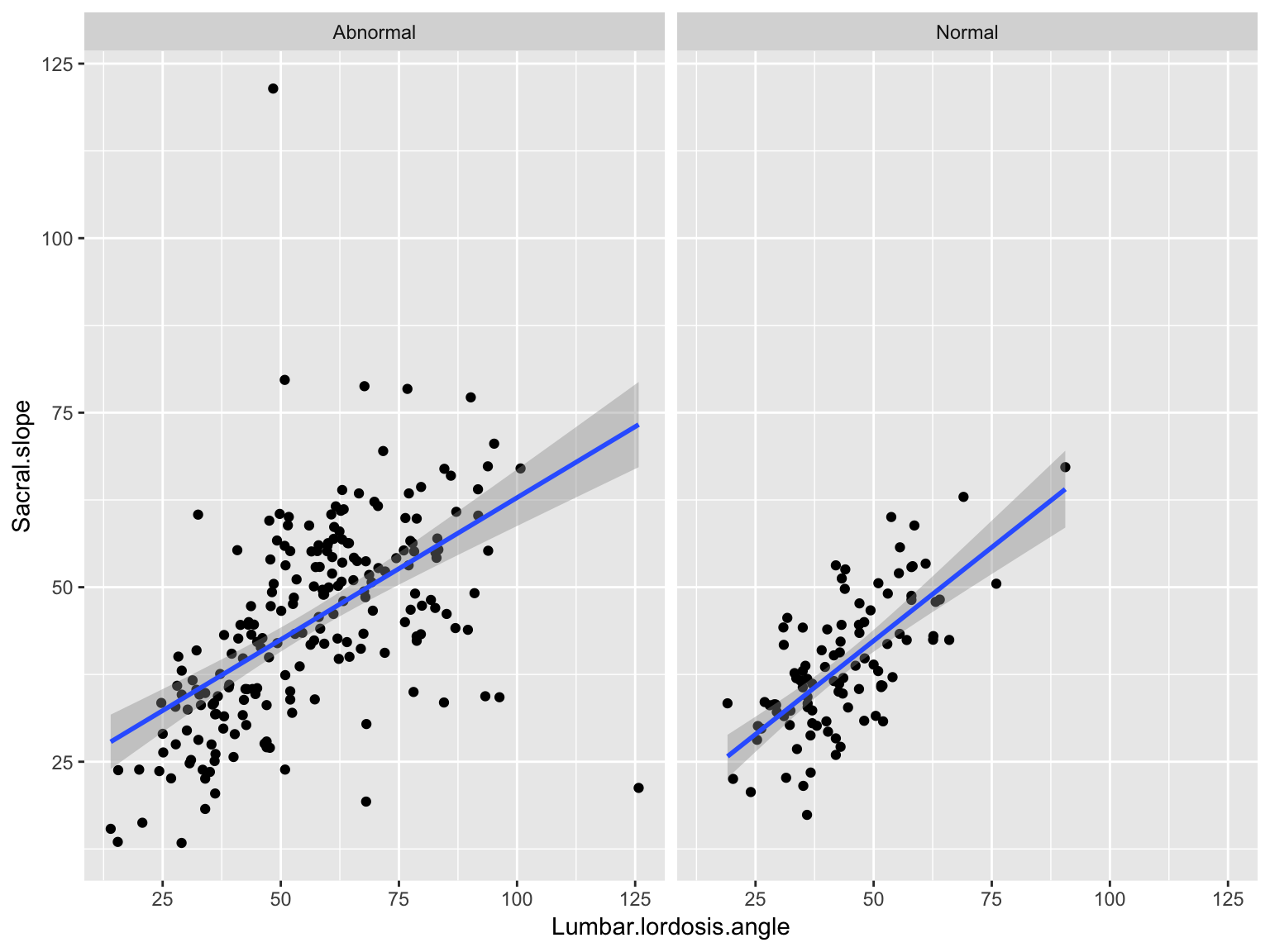
Manhattan plot
Manhattan plot
library(qqman)
data(gwasResults)
head(gwasResults)# SNP CHR BP P
# 1 rs1 1 1 0.9148060
# 2 rs2 1 2 0.9370754
# 3 rs3 1 3 0.2861395
# 4 rs4 1 4 0.8304476
# 5 rs5 1 5 0.6417455
# 6 rs6 1 6 0.5190959gwasResults <- gwasResults %>%
mutate(x_position = 1:n())Manhattan plot
ggplot(gwasResults, aes(x = x_position, y = -log(P, base=10)))+
geom_point(size = 0.2)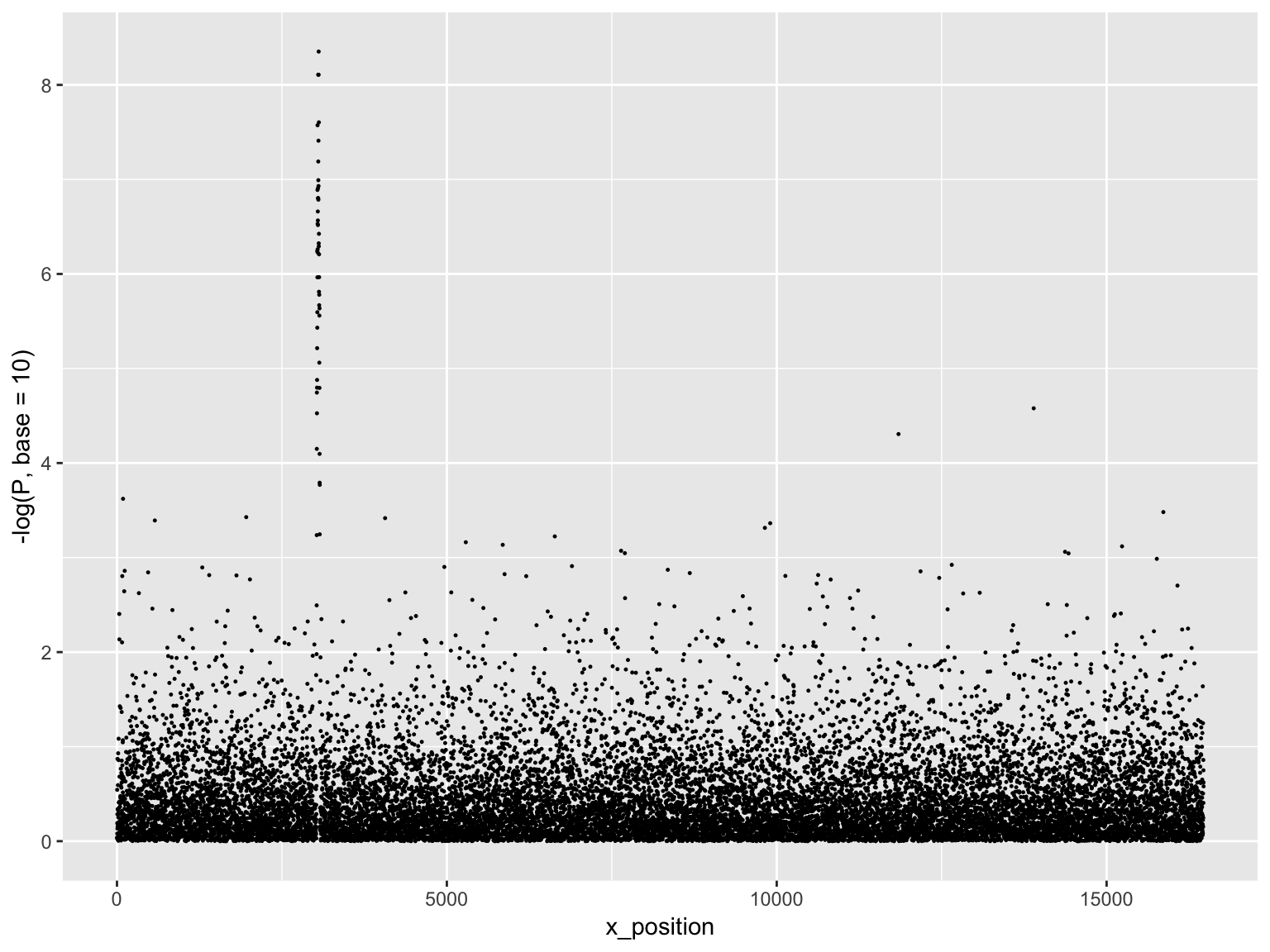
Manhattan plot
ggplot(gwasResults, aes(x = x_position, y = -log(P, base=10),
group=CHR, color=CHR))+
geom_point(size=0.2)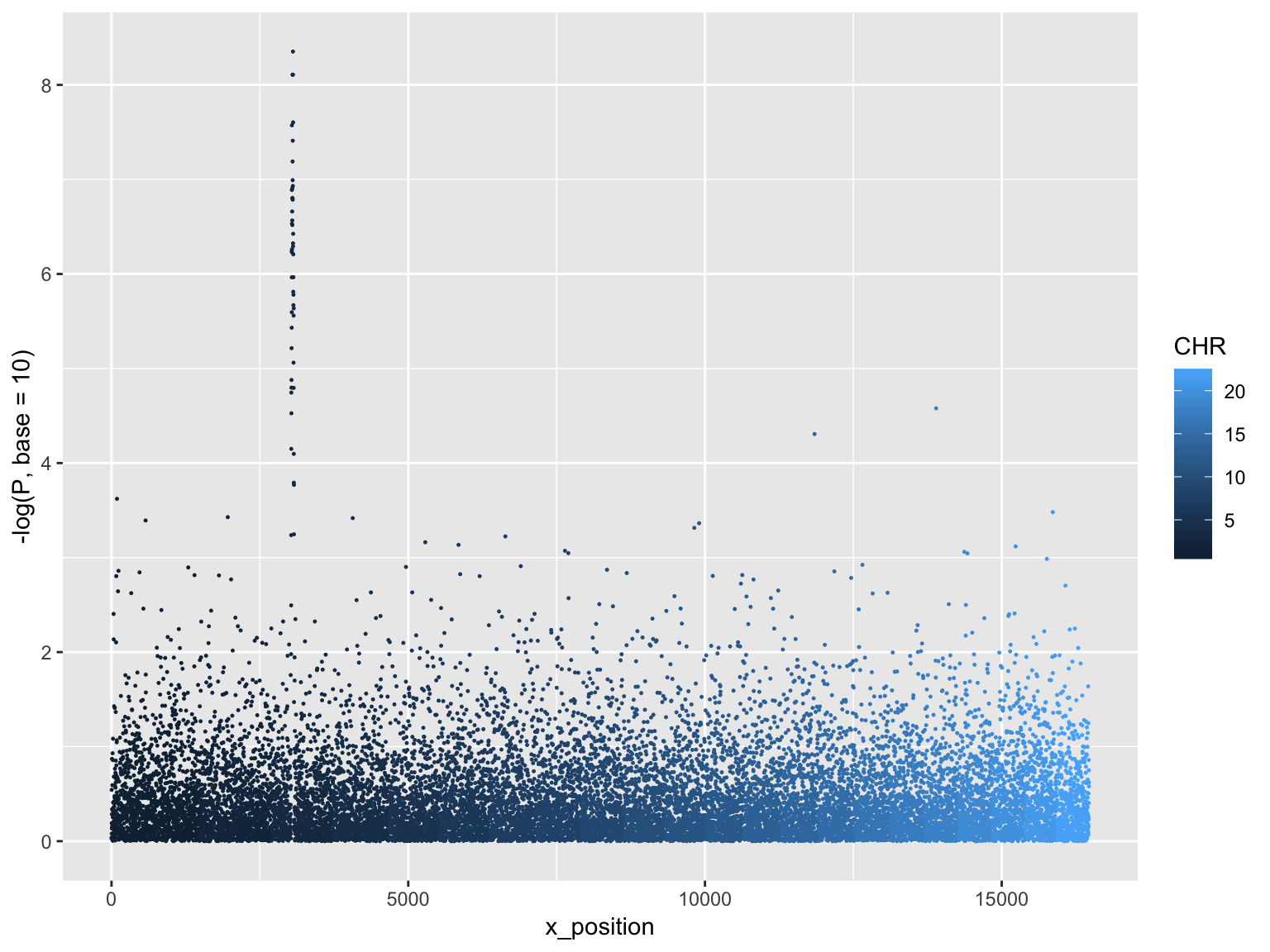
Manhattan plot
ggplot(gwasResults, aes(x = x_position, y = -log(P, base=10),
group=factor(CHR), color=factor(CHR)))+
geom_point(size=0.2)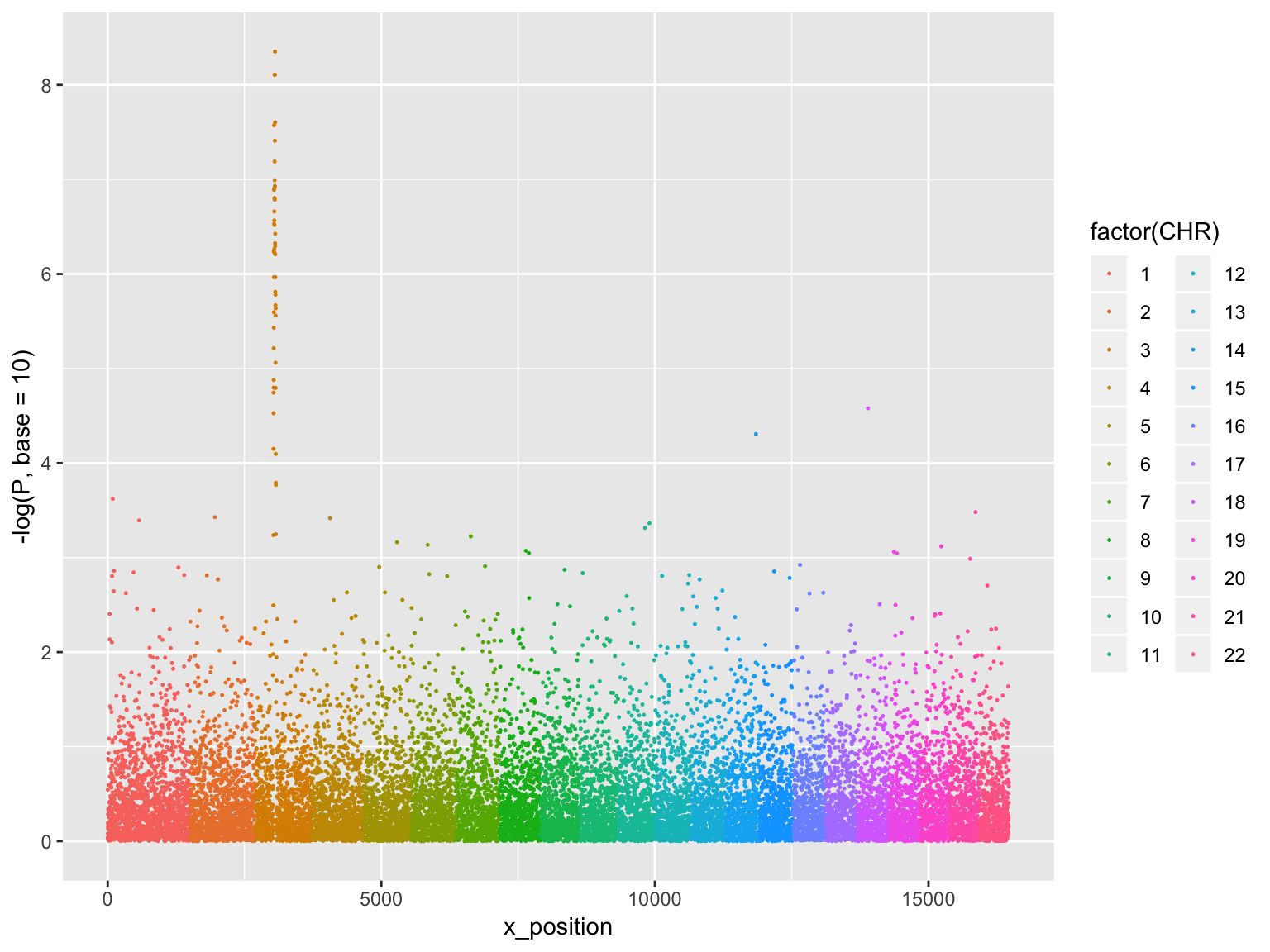
Manhattan plot
ggplot(gwasResults, aes(x = x_position, y = -log(P, base=10),
group=factor(CHR), color=factor(CHR)))+
geom_point(size=0.2)+
geom_hline(yintercept = 8, color='red', linetype=2)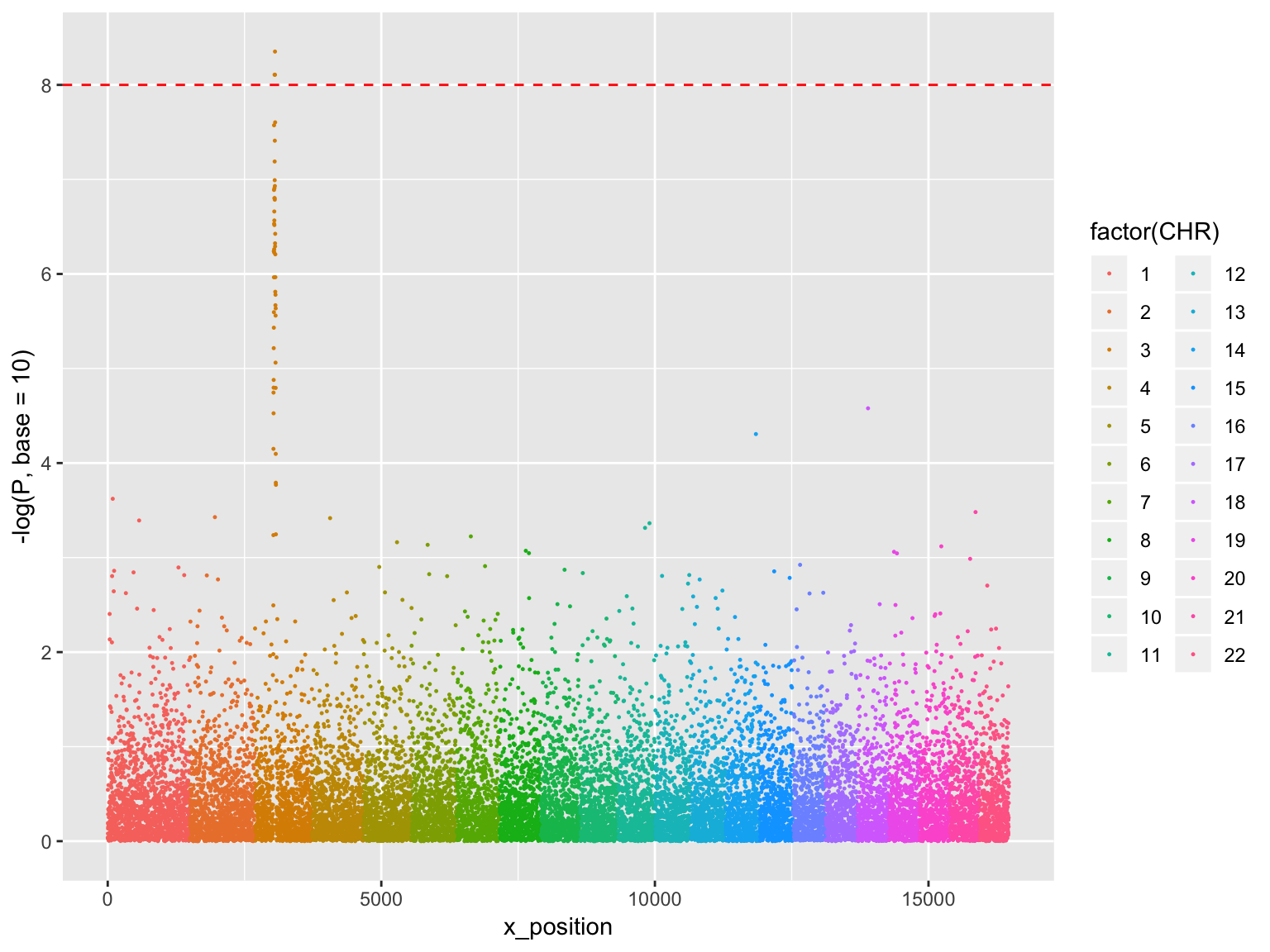
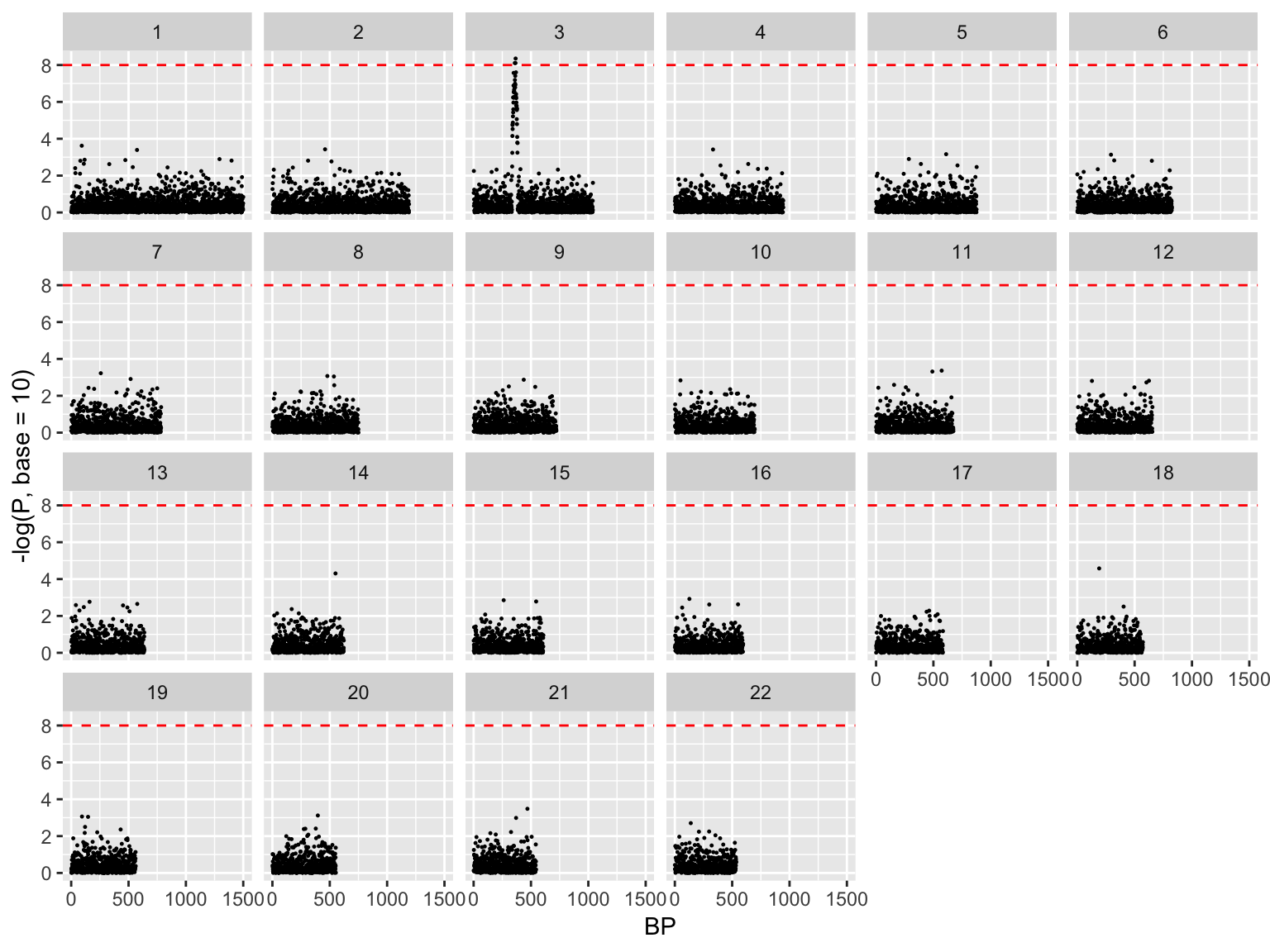
Manhattan plot, exploded
ggplot(gwasResults, aes(x = BP, y = -log(P, base=10)))+
geom_point(size=0.2)+
facet_wrap(~ CHR, nrow=4)+
geom_hline(yintercept = 8, color='red', linetype=2)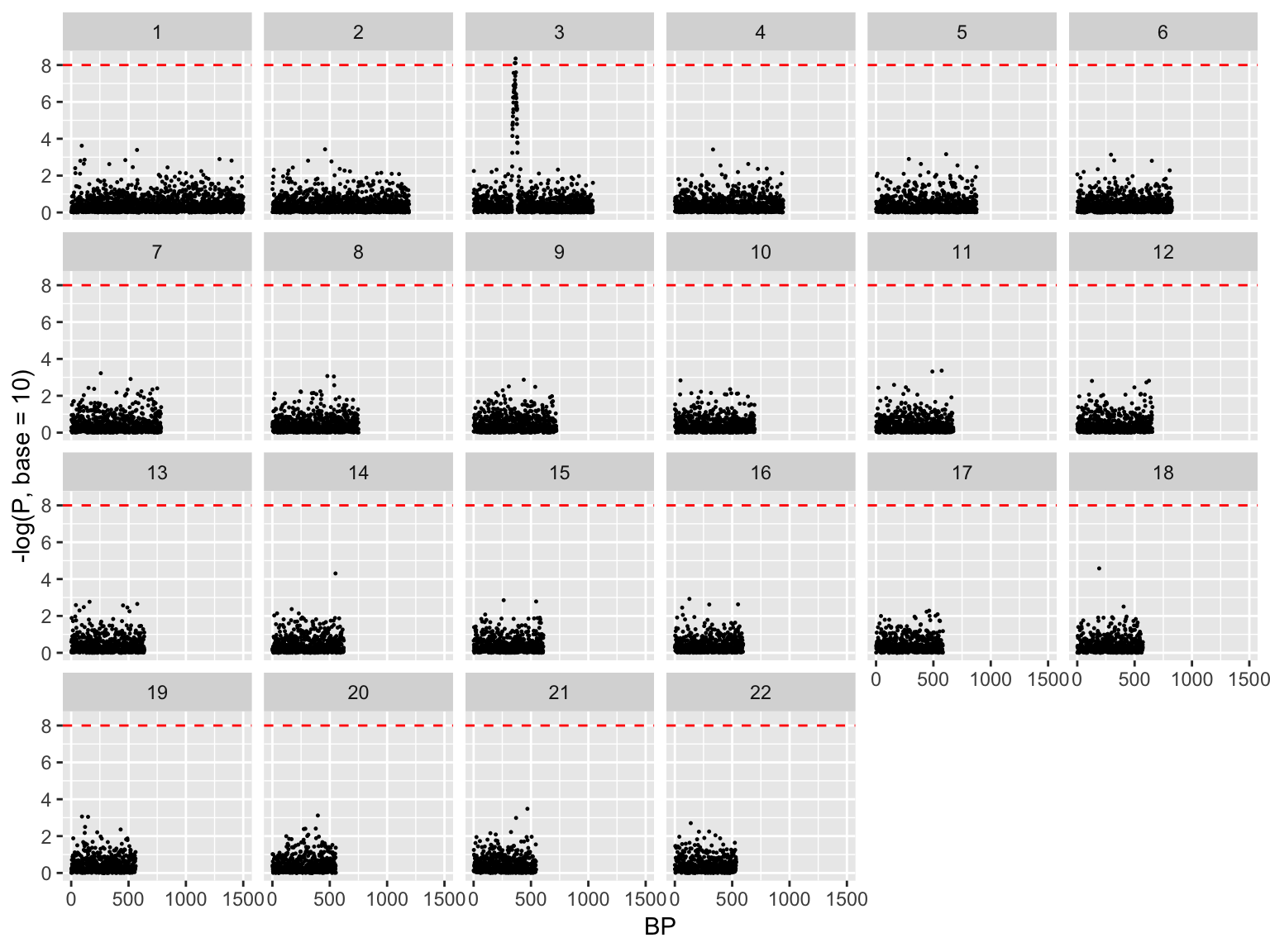
Manhattan plot, exploded